Final models evaluation
R For SAS Users

Melinda Higgins, PhD
Research Professor/Senior Biostatistician Emory University
Final exercises
Course wrap-up
Run regression models
- for different predictors
- for different groups
Choose best models
- save model results
- extract and display fit statistics
Showcase your skills
Evaluate and compare models
- using graphical visualizations
Report best associations
- between variables
- overall and by group
Comparing models
# Run lm() for diffht by bmi, save model
lmdiffhtbmi <- lm(diffht ~ bmi,
data = daviskeep)
# Run lm() for diffht by weight, save model
lmdiffhtwt <- lm(diffht ~ weight,
data = daviskeep)
# Run summary() for each model, save results
smrylmdiffhtbmi <- summary(lmdiffhtbmi)
smrylmdiffhtwt <- summary(lmdiffhtwt)
Comparing models
# Display r.squared for weight model
smrylmdiffhtwt$r.squared
# Display r.squared for bmi model
smrylmdiffhtbmi$r.squared
# Compare AICs for both models
AIC(lmdiffhtbmi, lmdiffhtwt)
[1] 0.003281645
[1] 0.00121824
df AIC
lmdiffhtbmi 3 788.0816
lmdiffhtwt 3 787.7052
Models by group - men vs women
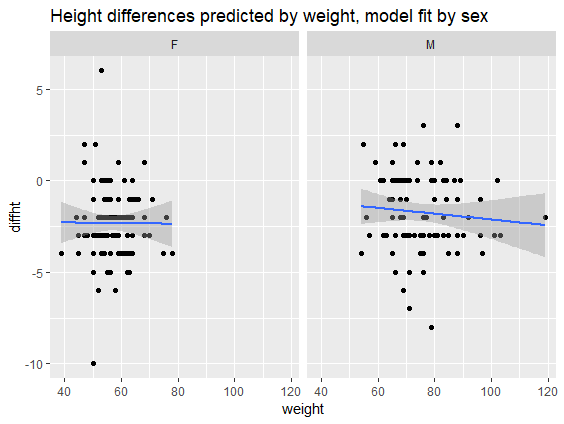
# Plot diffht by weight by sex
ggplot(daviskeep,
aes(diffht, weight)) +
geom_point() +
geom_smooth(method = "lm") +
facet_wrap(vars(sex)) +
ggtitle("Height differences
predicted by weight,
model fit by sex")
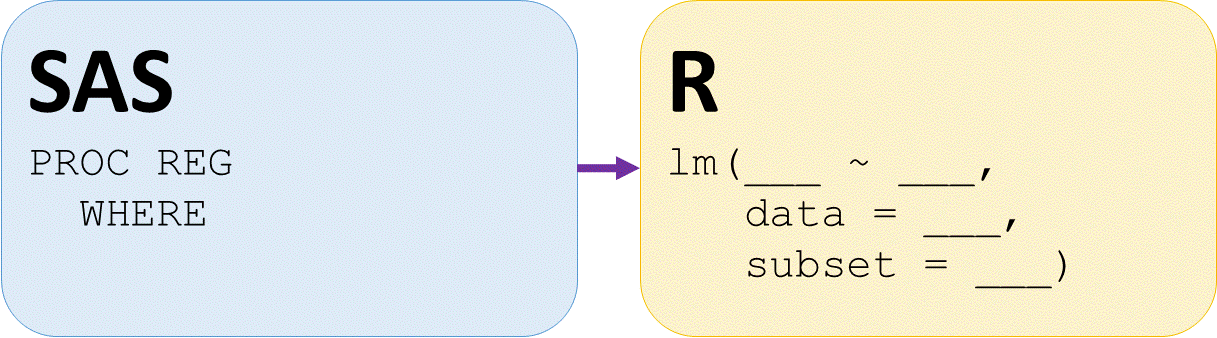
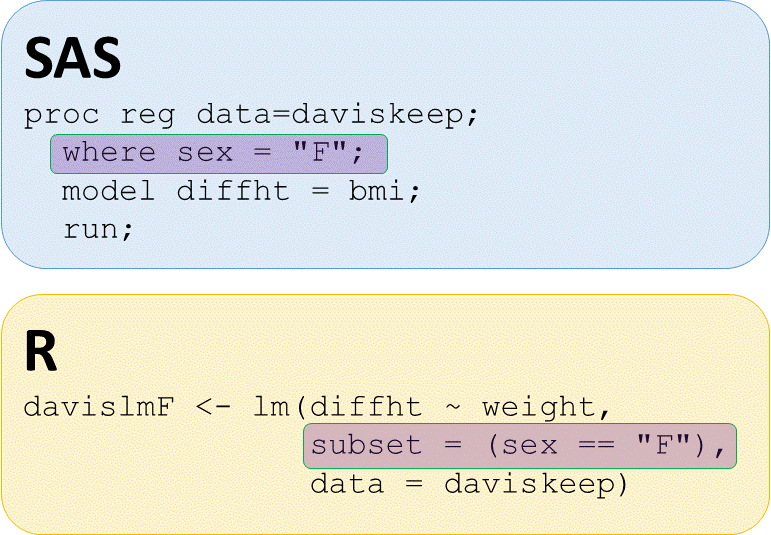
Regression on subset
Fit models for subsets
# lm() of diffht by weight for females
lmdiffhtwtF <- lm(diffht ~ weight,
subset = (sex == "F"),
data = daviskeep)
# lm() of diffht by weight for males
lmdiffhtwtM <- lm(diffht ~ weight,
subset = (sex == "M"),
data = daviskeep)
# Run summary() for each model save results
smrylmdiffhtwtF <- summary(lmdiffhtwtF)
smrylmdiffhtwtM <- summary(lmdiffhtwtM)
Fit models for subsets
# r.squared for females only model
smrylmdiffhtwtF$r.squared
# r.squared for males only model
smrylmdiffhtwtM$r.squared
[1] 4.00807e-05
[1] 0.00804139
Let's wrap up by developing a few models for predicting abalone ages!
R For SAS Users

