Data quality and cleaning
R For SAS Users

Melinda Higgins, PhD
Research Professor/Senior Biostatistician Emory University
Check distributions
# Continue with davismod
davismod %>%
head(5)
sex weight height repwt repht bmi diffht difflow bmicat
1 M 77 182 77 180 23.24598 -2 FALSE 1. underwt/norm
2 F 58 161 51 159 22.37568 -2 FALSE 1. underwt/norm
3 F 53 161 54 158 20.44674 -3 TRUE 1. underwt/norm
4 M 68 177 70 175 21.70513 -2 FALSE 1. underwt/norm
5 F 59 157 59 155 23.93606 -2 FALSE 1. underwt/norm
Check distributions
# Get summary statistics for bmi, check min, max, median
davismod %>%
pull(bmi) %>%
summary()
Min. 1st Qu. Median Mean 3rd Qu. Max.
15.82 20.23 21.84 24.70 23.94 510.93
Notice that Max. > 500
Visualize distributions
# Make bmi dotplot with geom_dotplot()
ggplot(davismod, aes(bmi)) +
geom_dotplot()
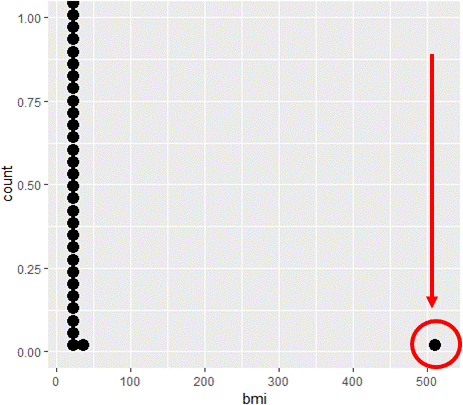
Find the outliers
# Sort data use arrange(), view last 6 rows with tail()
davismod %>%
arrange(bmi) %>%
tail()
sex weight height repwt repht bmi diffht difflow bmicat
195 M 89 173 86 173 29.73704 0 FALSE 2. overwt
196 M 102 185 107 185 29.80278 0 FALSE 2. overwt
197 M 103 185 101 182 30.09496 -3 TRUE 3. obese
198 M 101 183 100 180 30.15916 -3 TRUE 3. obese
199 M 119 180 124 178 36.72840 -2 FALSE 3. obese
200 F 166 57 56 163 510.92644 106 FALSE 3. obese
Visualize assumption that weight <= height
# Scatterplot with y=x reference line
ggplot(davismod,
aes(weight, height)) +
geom_point() +
geom_abline(intercept=0, slope=1)
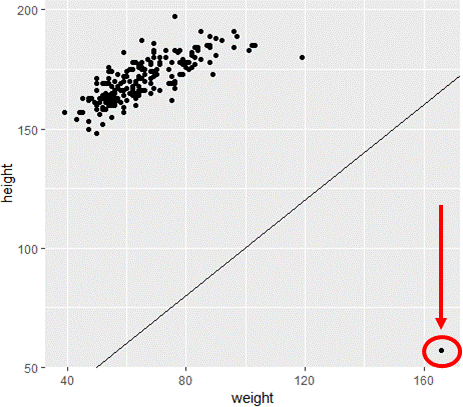
Filter out cases with errors
# Use filter() from dplyr, keep cases for bmi < 100
daviskeep <- davismod %>%
filter(bmi < 100)
# View last 6 rows
daviskeep %>%
arrange(bmi) %>%
tail()
sex weight height repwt repht bmi diffht difflow bmicat
194 F 75 162 75 158 28.57796 -4 TRUE 2. overwt
195 M 89 173 86 173 29.73704 0 FALSE 2. overwt
196 M 102 185 107 185 29.80278 0 FALSE 2. overwt
197 M 103 185 101 182 30.09496 -3 TRUE 3. obese
198 M 101 183 100 180 30.15916 -3 TRUE 3. obese
199 M 119 180 124 178 36.72840 -2 FALSE 3. obese
Visualize corrected bmi
# Make dotplot of bmi
ggplot(daviskeep, aes(bmi)) +
geom_dotplot()
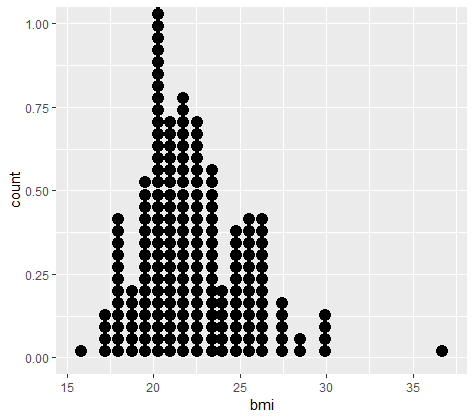
Final cleanup of abalone dataset
- Check the assumptions of the abalone dataset
- Remove cases that violate assumptions
- Finalize dataset for analysis and models
ASSUMPTIONS:
- All measurements should be > 0
lengthis the longest shell dimensionheightanddiameter<lengthwholeWeightis the total weight- Other weights <
wholeWeight

Let's explore and clean up the abalone dataset
R For SAS Users

