ANOVA and linear models
R For SAS Users

Melinda Higgins, PhD
Research Professor/Senior Biostatistician Emory University
Note on missing values
# Stats by bmicat - descriptives for ANOVA
daviskeep %>%
group_by(bmicat) %>%
select(diffht, bmicat) %>%
summarise(across(everything(),
list(mean = ~mean(.x),
sd = ~sd(.x),
var = ~var(.x))),
N = n())
# A tibble: 3 × 5
bmicat diffht_mean diffht_sd diffht_var N
<chr> <dbl> <dbl> <dbl> <int>
1 1. underwt/norm NA NA NA 161
2 2. overwt NA NA NA 35
3 3. obese -2.67 0.577 0.333 3
Note on missing values
# Stats by bmicat - descriptives for ANOVA
daviskeep %>%
group_by(bmicat) %>%
select(diffht, bmicat) %>%
summarise(across(everything(),
list(mean = ~mean(.x, na.rm = TRUE),
sd = ~sd(.x, na.rm = TRUE),
var = ~var(.x, na.rm = TRUE))),
N = n())
# A tibble: 3 × 5
bmicat diffht_mean diffht_sd diffht_var N
<chr> <dbl> <dbl> <dbl> <int>
1 1. underwt/norm -2.12 2.14 4.56 161
2 2. overwt -1.78 1.91 3.66 35
3 3. obese -2.67 0.577 0.333 3
Analysis of Variance (ANOVA) SAS and R
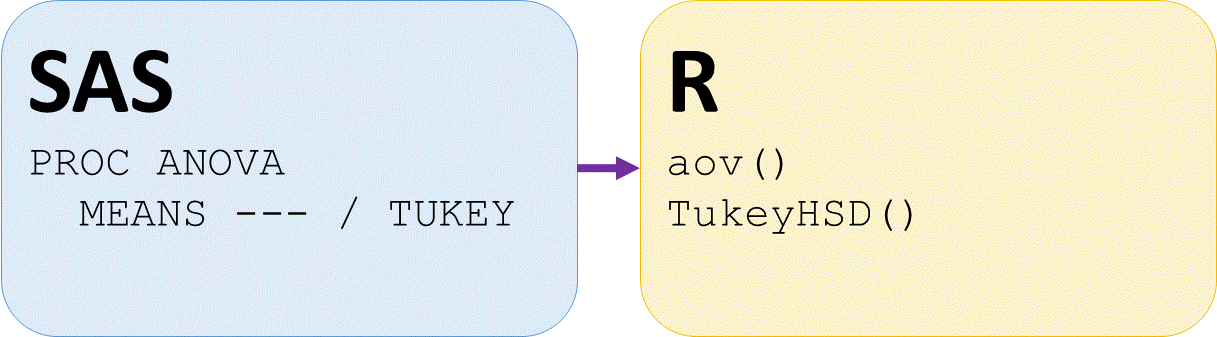
Analysis of Variance (ANOVA)
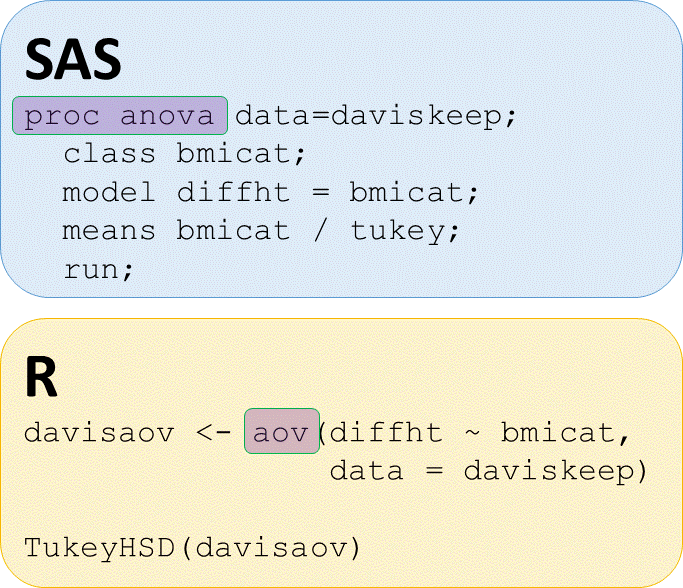
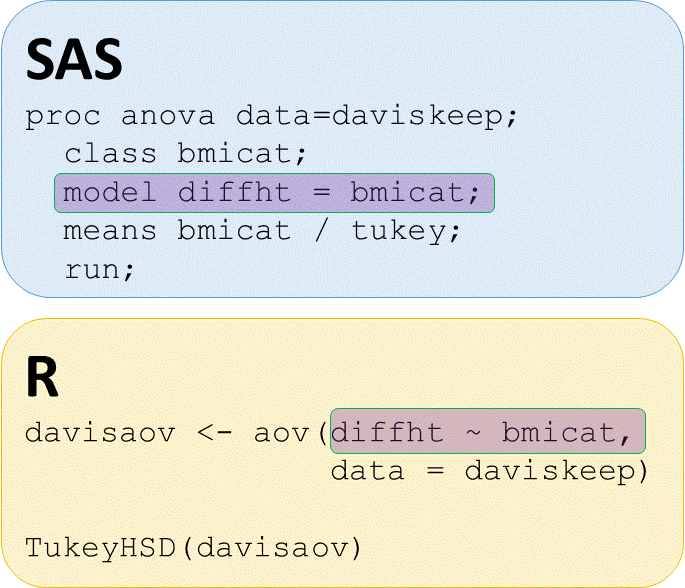
# Perform ANOVA of diffht by bmicat, save output as davisaov
davisaov <- aov(diffht ~ bmicat, data = daviskeep)
# Show summary of davisaov
summary(davisaov)
Df Sum Sq Mean Sq F value Pr(>F)
bmicat 2 4.1 2.070 0.475 0.623
Residuals 179 779.9 4.357
17 observations deleted due to missingness
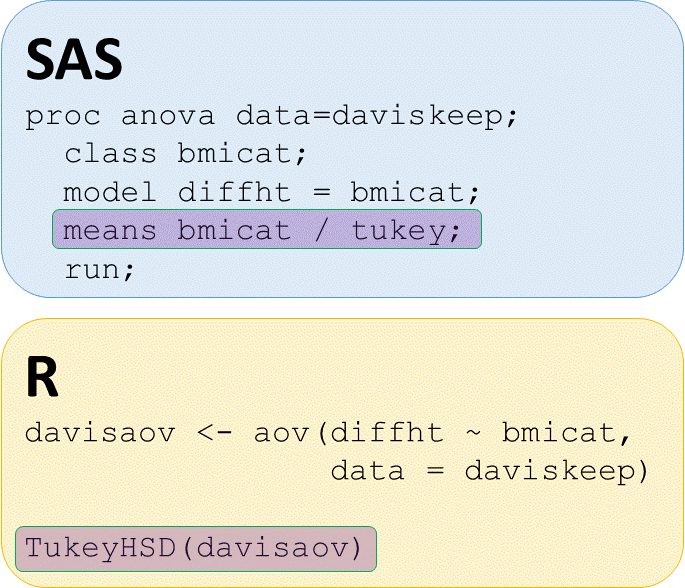
Post hoc pairwise tests
# Perform TukeyHSD posthoc pairwise tests on abaov
TukeyHSD(davisaov)
Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = diffht ~ bmicat, data = daviskeep)
$bmicat
diff lwr upr p adj
2. overwt-1. underwt/norm 0.3411990 -0.6211376 1.303536 0.6799698
3. obese-1. underwt/norm -0.5442177 -3.4213543 2.332919 0.8957761
3. obese-2. overwt -0.8854167 -3.8641564 2.093323 0.7623194
Linear regression SAS and R
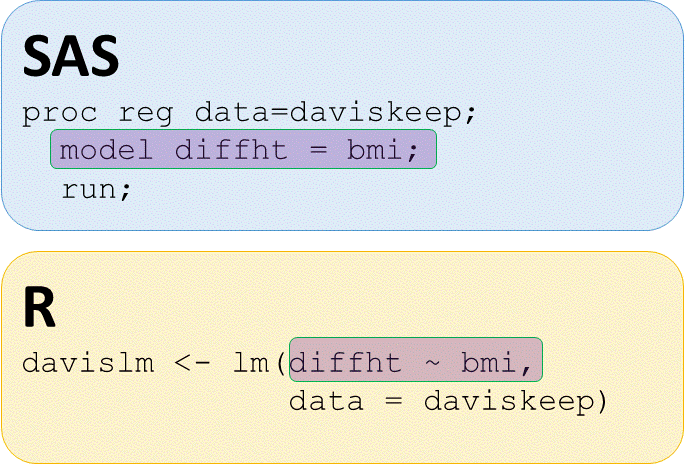
Simple linear regression
# Run lm() of diffht by bmi
davislm <- lm(diffht ~ bmi,
data = daviskeep)
davislm
# Display elements in davislm
names(davislm)
Call:
lm(formula = diffht ~ bmi,
data = daviskeep)
Coefficients:
(Intercept) bmi
-2.60878 0.02404
[1] "coefficients" "residuals"
[3] "effects" "rank"
[5] "fitted.values" "assign"
[7] "qr" "df.residual"
[9] "na.action" "xlevels"
[11] "call" "terms"
[13] "model"
Simple linear regression
# Display coefficients element
davislm$coefficients
(Intercept) bmi
-2.60877657 0.02403939
# Display the slope coefficient 2
davislm$coefficients[2]
bmi
0.02403939
summary(davislm)
Call:
lm(formula = diffht ~ bmi, data = daviskeep)
Residuals:
Min 1Q Median 3Q Max
-7.8607 -0.9944 -0.0048 1.1271 8.1627
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -2.60878 1.15718 -2.254 0.0254 *
bmi 0.02404 0.05130 0.469 0.6399
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 2.086 on 180 degrees of freedom
(17 observations deleted due to missingness)
Multiple R-squared: 0.001218, Adjusted R-squared: -0.004331
F-statistic: 0.2196 on 1 and 180 DF, p-value: 0.6399
Summary of linear regression model
# Save summary() output of lmshucked, see element names
smrydavislm <- summary(davislm)
names(smrydavislm)
[1] "call" "terms" "residuals" "coefficients"
[5] "aliased" "sigma" "df" "r.squared"
[9] "adj.r.squared" "fstatistic" "cov.unscaled" "na.action"
Summary of linear regression model
# Display r.squared from smrydavislm
smrydavislm$r.squared
[1] 0.00121824
# Display coefficients from smrydavislm
smrydavislm$coefficients
Estimate Std. Error t value Pr(>|t|)
(Intercept) -2.60877657 1.15717685 -2.2544320 0.02537425
bmi 0.02403939 0.05130456 0.4685624 0.63994943
Let's go fit and explore models for abalones!
R For SAS Users

