Categorical data: analyze and visualize
R For SAS Users

Melinda Higgins, PhD
Research Professor/Senior Biostatistician Emory University
Collapse categories
# Use table() inside with() for bmicat
daviskeep %>% with(table(bmicat))
bmicat
1. underwt/norm 2. overwt 3. obese
161 35 3
Add recoded variable bmigt25
# Add one more categorical variable bmigt25
daviskeep <- daviskeep %>%
mutate(bmigt25 = ifelse(bmi > 25,
"2. overwt/obese",
"1. underwt/norm"))
# View frequencies for bmigt25 categories
daviskeep %>% with(table(bmigt25))
bmigt25
1. underwt/norm 2. overwt/obese
161 38
Contingency tables SAS and R
Chi-square tests SAS and R
Contingency table and chi-square test
# Save table output of bmigt25 by sex
tablebmisex <- daviskeep %>%
with(table(bmigt25, sex))
tablebmisex
# Use table object to run chisq.test
chisq.test(tablebmisex)
sex
bmigt25 F M
1. underwt/norm 107 54
2. overwt/obese 4 34
Pearson's Chi-squared test with Yates'
continuity correction
data: tablebmisex
X-squared = 36.759, df = 1, p-value = 1.336e-09
Chi-square tests with gmodels package
# Load gmodel package
library(gmodels)
# Run gmodels::CrossTabs, show column %s and expected values
daviskeep %>%
with(gmodels::CrossTable(bmigt25, sex,
chisq = TRUE,
prop.r = FALSE,
prop.t = FALSE,
prop.chisq = FALSE,
expected = TRUE))
CrossTable output - part 1
Cell Contents
|-------------------------|
| N |
| Expected N |
| N / Col Total |
|-------------------------|
Total Observations in Table: 199
| sex
bmigt25 | F | M | Row Total |
----------------|-----------|-----------|-----------|
1. underwt/norm | 107 | 54 | 161 |
| 89.804 | 71.196 | |
| 0.964 | 0.614 | |
----------------|-----------|-----------|-----------|
2. overwt/obese | 4 | 34 | 38 |
| 21.196 | 16.804 | |
| 0.036 | 0.386 | |
----------------|-----------|-----------|-----------|
Column Total | 111 | 88 | 199 |
| 0.558 | 0.442 | |
----------------|-----------|-----------|-----------|
CrossTable output - part 2
gmodels::CrossTable() output - continued...
Statistics for All Table Factors
Pearson's Chi-squared test
------------------------------------------------------------
Chi^2 = 38.99402 d.f. = 1 p = 4.251066e-10
Pearson's Chi-squared test with Yates' continuity correction
------------------------------------------------------------
Chi^2 = 36.75936 d.f. = 1 p = 1.336475e-09
Mosaic plots SAS and R
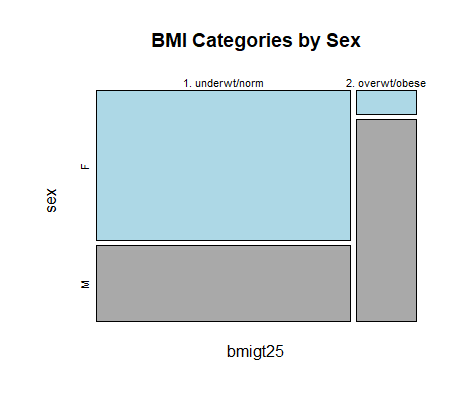
Mosaicplot of two-way categorical proportions
# Make mosaicplot of bmigt25 by sex
mosaicplot(bmigt25 ~ sex,
data = daviskeep,
color = c("light blue",
"dark grey"),
main =
"BMI Categories by Sex")
Let's explore categorical associations for the abalones!
R For SAS Users

