Principal component analysis
Dimensionality Reduction in Python

Jeroen Boeye
Head of Machine Learning, Faktion
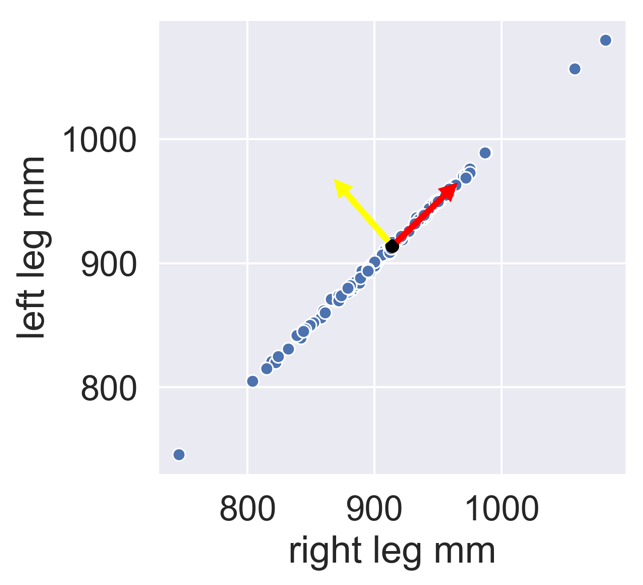
PCA concept
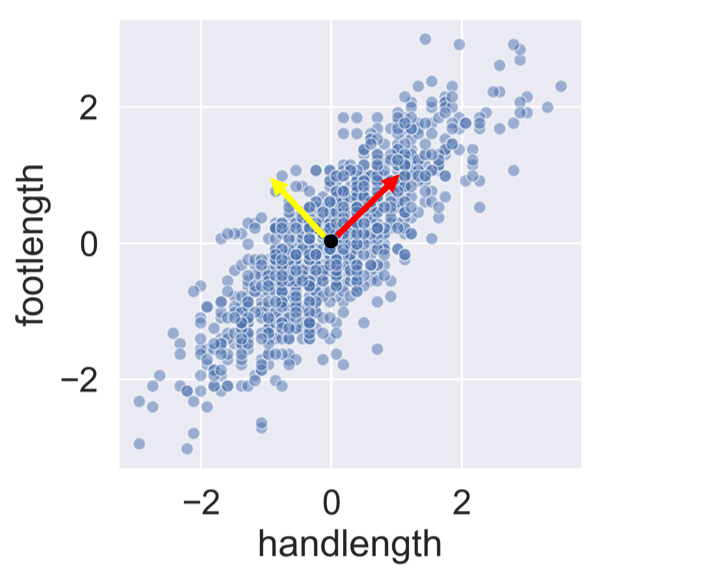
PCA concept
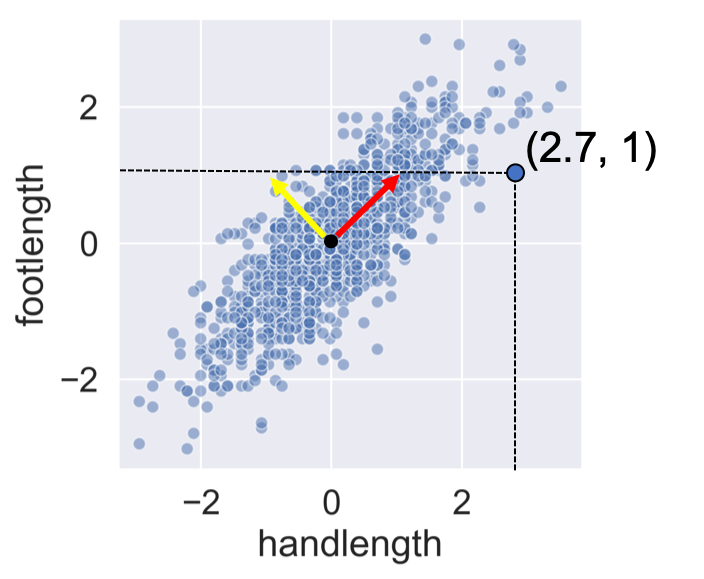
PCA concept
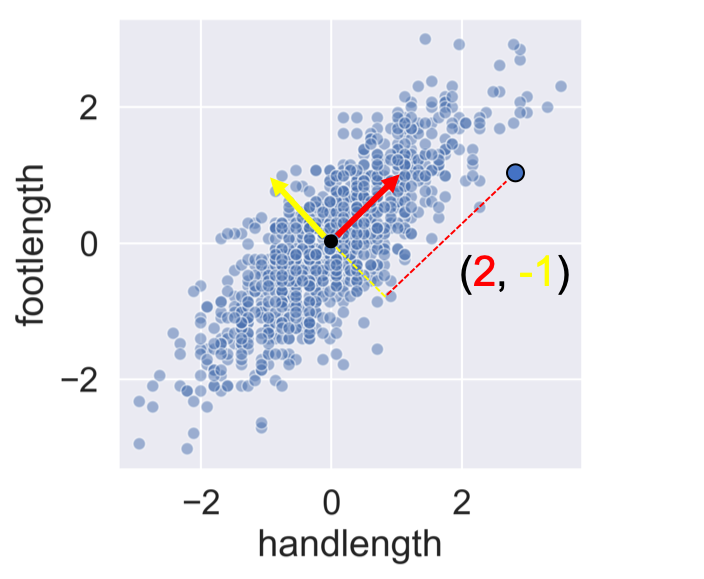
Calculating the principal components
from sklearn.preprocessing import StandardScaler scaler = StandardScaler() std_df = scaler.fit_transform(df)from sklearn.decomposition import PCA pca = PCA() print(pca.fit_transform(std_df))
[[-0.08320426 -0.12242952]
[ 0.31478004 0.57048158]
...
[-0.5609523 0.13713944]
[-0.0448304 -0.37898246]]
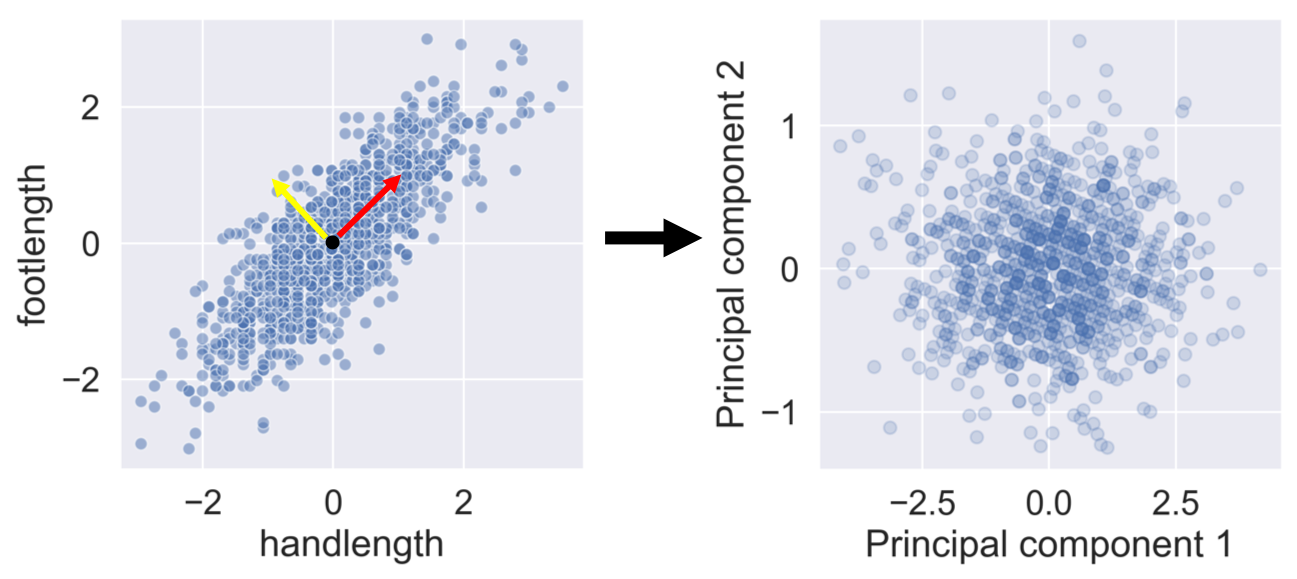
PCA removes correlation
Principal component explained variance ratio
from sklearn.decomposition import PCA
pca = PCA()
pca.fit(std_df)
print(pca.explained_variance_ratio_)
array([0.90, 0.10])
PCA for dimensionality reduction
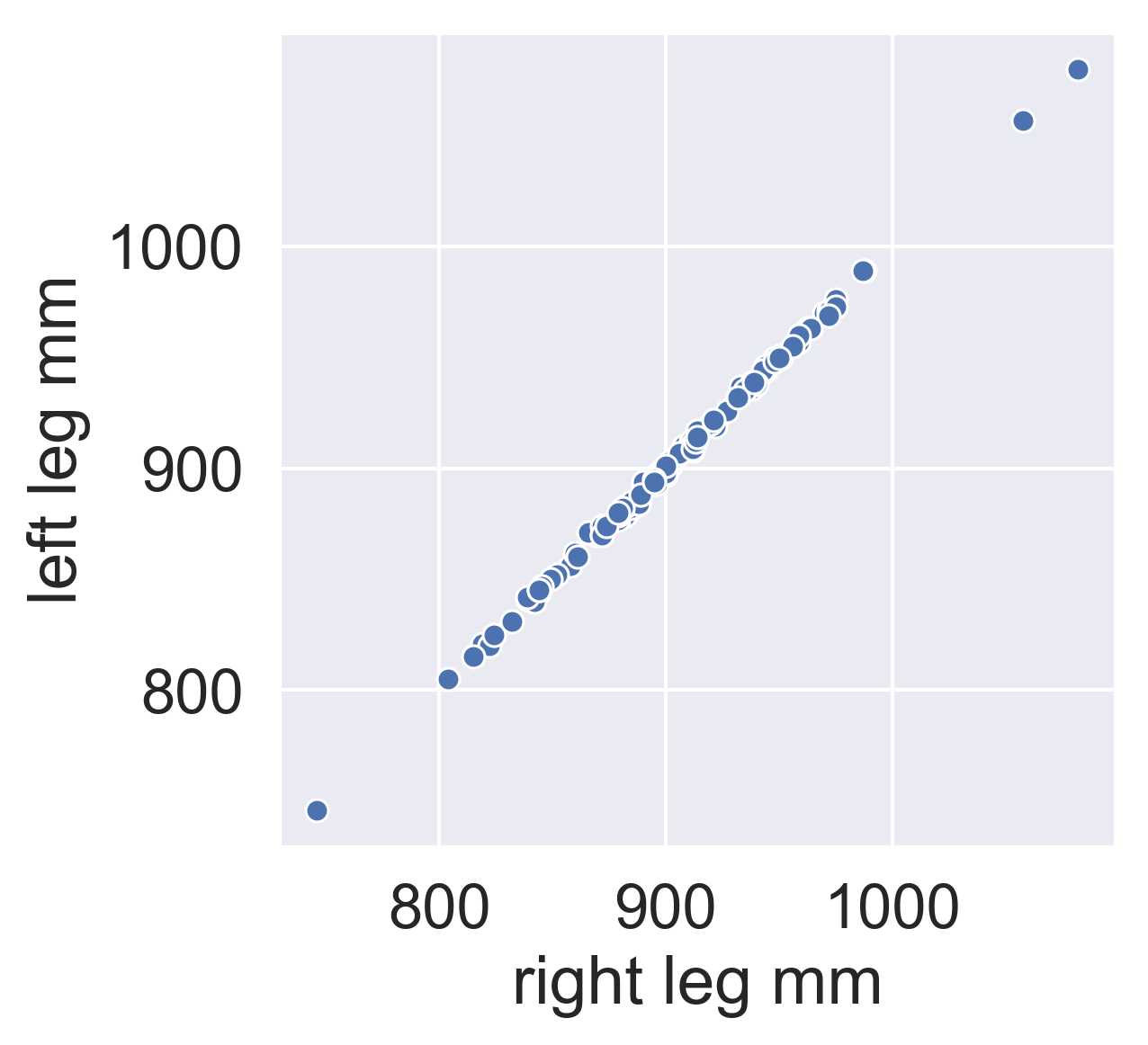
PCA for dimensionality reduction
print(pca.explained_variance_ratio_)
array([0.9997, 0.0003])
PCA for dimensionality reduction
pca = PCA()
pca.fit(ansur_std_df)
print(pca.explained_variance_ratio_)
array([0.44, 0.18, 0.04, 0.03, 0.02, 0.02, 0.02, 0.01, 0.01, 0.01, 0.01,
0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01,
0.01, 0.01, 0.01, 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
...
0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. , 0. ])
PCA for dimensionality reduction
pca = PCA()
pca.fit(ansur_std_df)
print(pca.explained_variance_ratio_.cumsum())
array([0.44, 0.62, 0.66, 0.69, 0.72, 0.74, 0.76, 0.77, 0.79, 0.8 , 0.81,
0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.87, 0.88, 0.89, 0.89, 0.9 ,
0.9 , 0.91, 0.92, 0.92, 0.92, 0.93, 0.93, 0.94, 0.94, 0.94, 0.95,
...
0.99, 0.99, 0.99, 0.99, 0.99, 1. , 1. , 1. , 1. , 1. , 1. ,
1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. , 1. ,
1. , 1. , 1. , 1. , 1. , 1. ])
Let's practice!
Dimensionality Reduction in Python

