Principal Component selection
Dimensionality Reduction in Python

Jeroen Boeye
Head of Machine Learning, Faktion
Setting an explained variance threshold
pipe = Pipeline([ ('scaler', StandardScaler()), ('reducer', PCA(n_components=0.9))])# Fit the pipe to the data pipe.fit(poke_df) print(len(pipe['reducer'].components_))
5
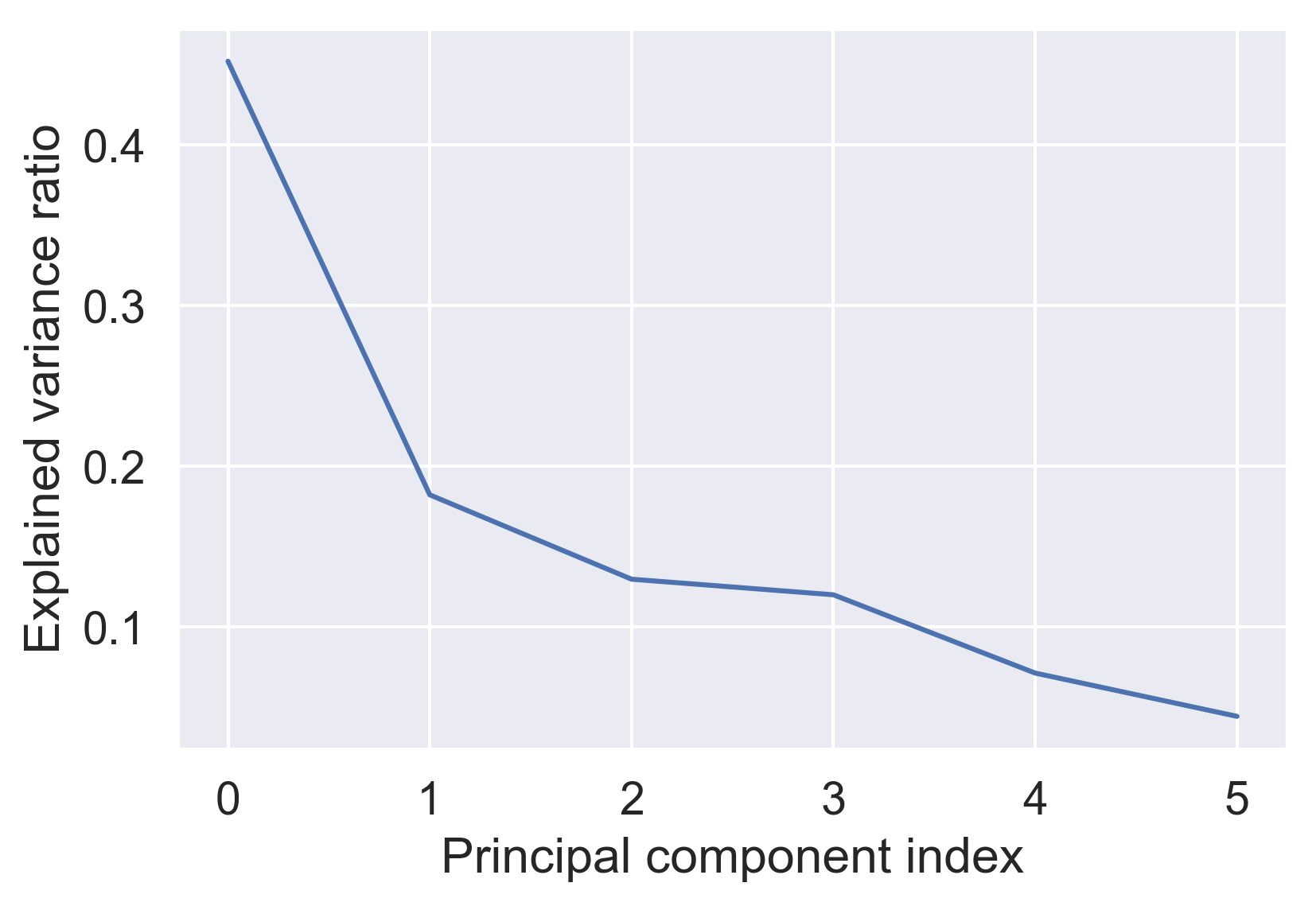
An optimal number of components
pipe.fit(poke_df)
var = pipe['reducer'].explained_variance_ratio_
plt.plot(var)
plt.xlabel('Principal component index')
plt.ylabel('Explained variance ratio')
plt.show()
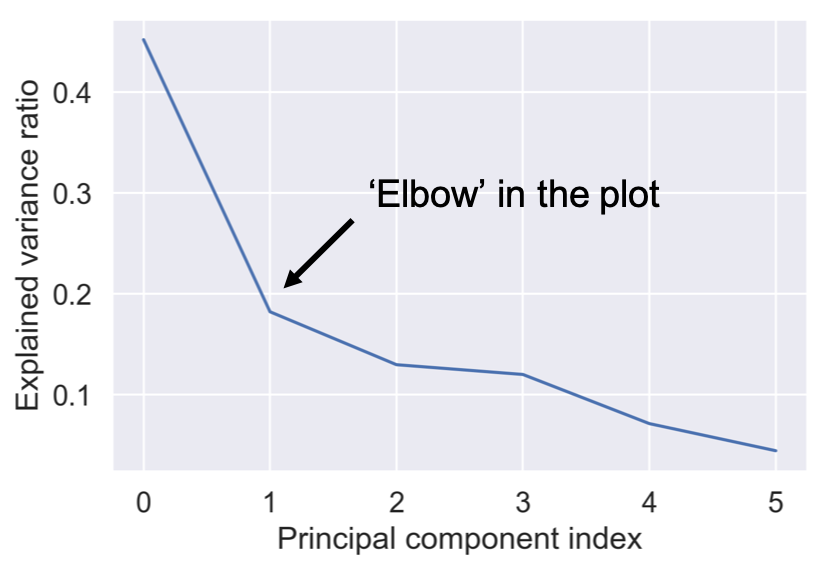
An optimal number of components
pipe.fit(poke_df)
var = pipe['reducer'].explained_variance_ratio_
plt.plot(var)
plt.xlabel('Principal component index')
plt.ylabel('Explained variance ratio')
plt.show()
PCA operations

PCA operations

PCA operations

Compressing images
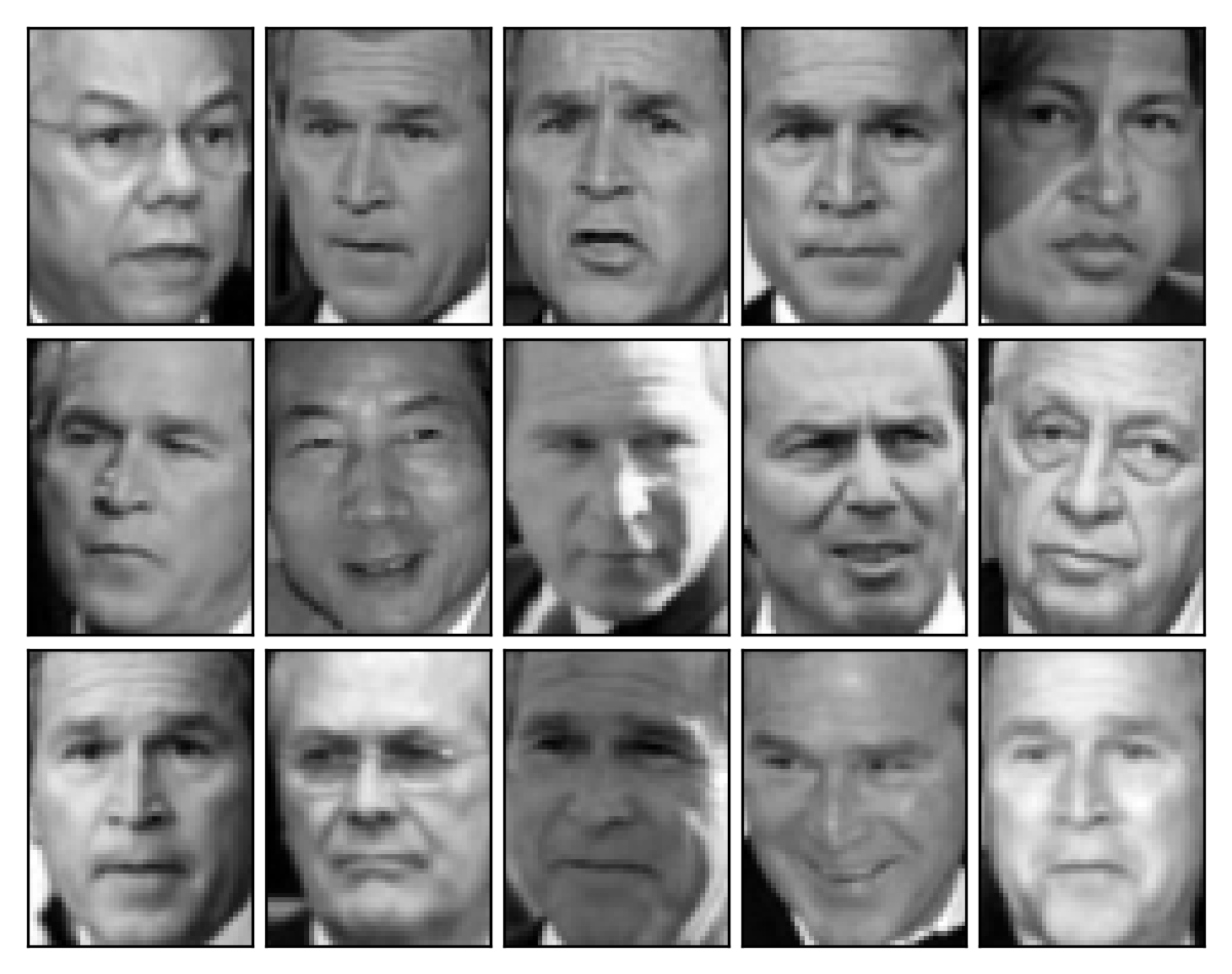
Compressing images
print(X_test.shape)
(15, 2914)
62 x 47 pixels = 2914 grayscale values
print(X_train.shape)
(1333, 2914)
Compressing images
pipe = Pipeline([ ('scaler', StandardScaler()), ('reducer', PCA(n_components=290))])pipe.fit(X_train)pc = pipe.fit_transform(X_test) print(pc.shape)
(15, 290)
Rebuilding images
pc = pipe.transform(X_test)
print(pc.shape)
(15, 290)
X_rebuilt = pipe.inverse_transform(pc)
print(X_rebuilt.shape)
(15, 2914)
img_plotter(X_rebuilt)

Rebuilding images


Let's practice!
Dimensionality Reduction in Python

