PCA applications
Dimensionality Reduction in Python

Jeroen Boeye
Head of Machine Learning, Faktion
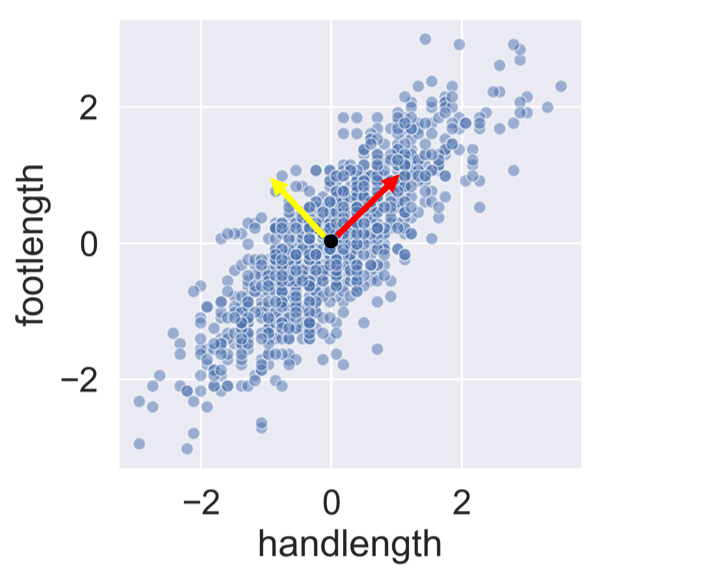
Understanding the components
print(pca.components_)
array([[ 0.71, 0.71],
[ -0.71, 0.71]])
PC 1 = 0.71 x Hand length + 0.71 x Foot length
PC 2 = -0.71 x Hand length + 0.71 x Foot length
PCA for data exploration
PCA in a pipeline
from sklearn.preprocessing import StandardScaler from sklearn.decomposition import PCA from sklearn.pipeline import Pipeline pipe = Pipeline([ ('scaler', StandardScaler()), ('reducer', PCA())])pc = pipe.fit_transform(ansur_df) print(pc[:,:2])
array([[-3.46114925, 1.5785215 ],
[ 0.90860615, 2.02379935],
...,
[10.7569818 , -1.40222755],
[ 7.64802025, 1.07406209]])
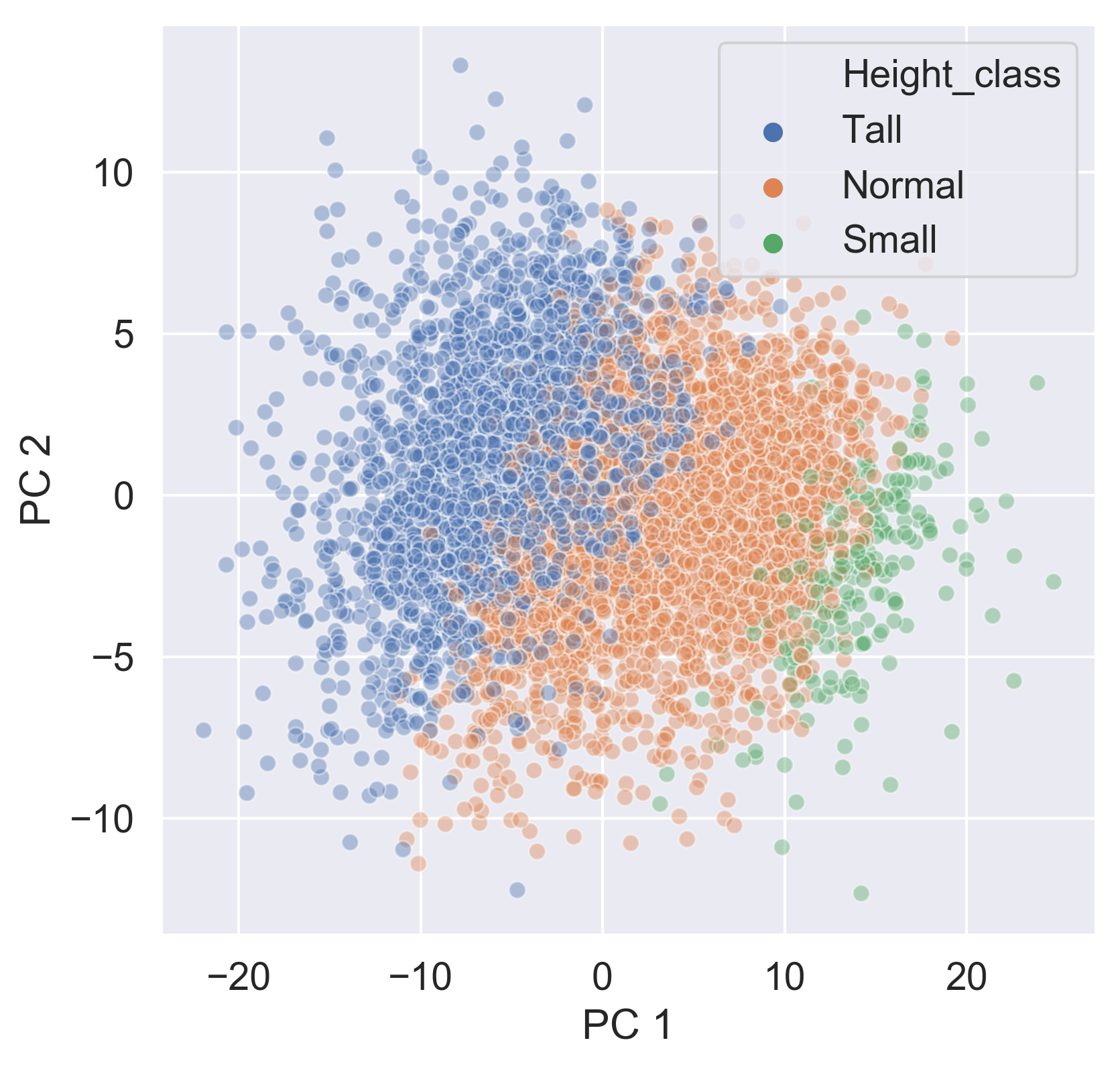
Checking the effect of categorical features
print(ansur_categories.head())
Branch Component Gender BMI_class Height_class
0 Combat Arms Regular Army Male Overweight Tall
1 Combat Support Regular Army Male Overweight Normal
2 Combat Support Regular Army Male Overweight Normal
3 Combat Service Support Regular Army Male Overweight Normal
4 Combat Service Support Regular Army Male Overweight Tall
Checking the effect of categorical features
ansur_categories['PC 1'] = pc[:,0] ansur_categories['PC 2'] = pc[:,1]sns.scatterplot(data=ansur_categories, x='PC 1', y='PC 2', hue='Height_class', alpha=0.4)

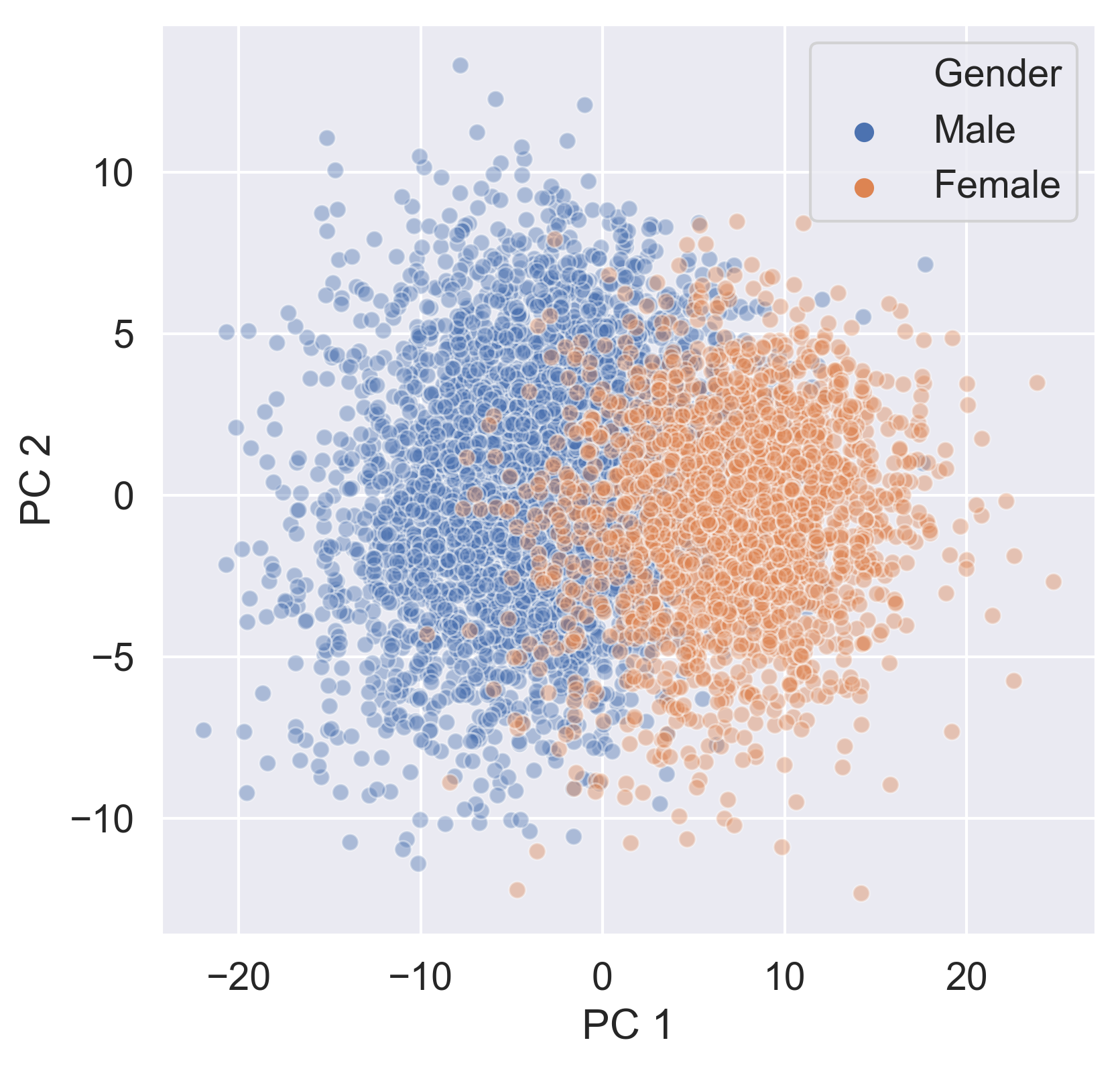
Checking the effect of categorical features
sns.scatterplot(data=ansur_categories,
x='PC 1', y='PC 2',
hue='Gender', alpha=0.4)
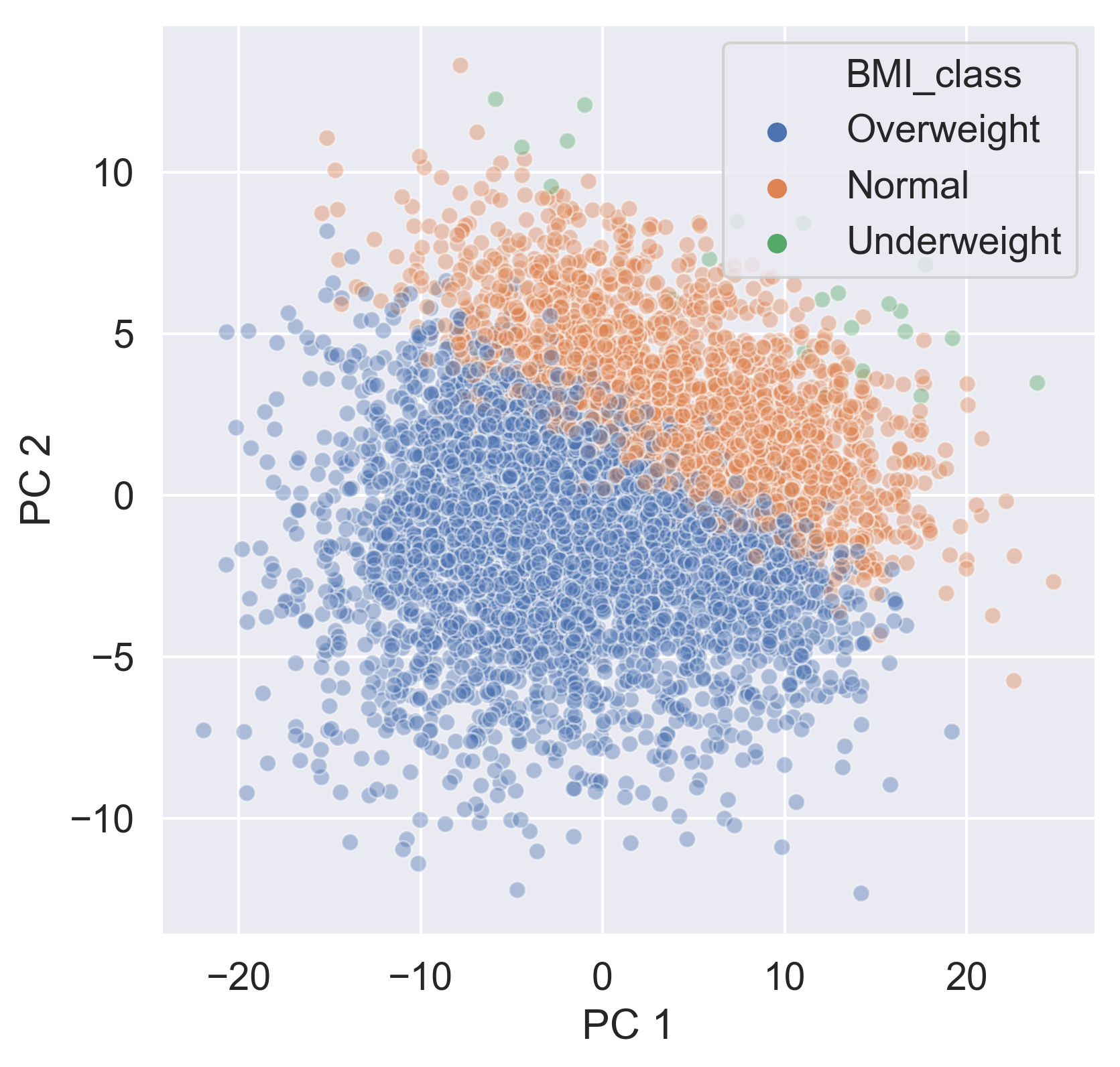
Checking the effect of categorical features
sns.scatterplot(data=ansur_categories,
x='PC 1', y='PC 2',
hue='BMI_class', alpha=0.4)
PCA in a model pipeline
pipe = Pipeline([ ('scaler', StandardScaler()), ('reducer', PCA(n_components=3)), ('classifier', RandomForestClassifier())])print(pipe['reducer'])
PCA(n_components=3)
PCA in a model pipeline
pipe.fit(X_train, y_train)pipe['reducer'].explained_variance_ratio_
array([0.56, 0.13, 0.05])
pipe['reducer'].explained_variance_ratio_.sum()
0.74
print(pipe.score(X_test, y_test))
0.986
Let's practice!
Dimensionality Reduction in Python

