Evaluation of different imputation techniques
Dealing with Missing Data in Python

Suraj Donthi
Deep Learning & Computer Vision Consultant
Evaluation techniques
- Imputations are used to improve model performance.
- Imputation with maximum machine learning model performance is selected.
- Density plots explain the distribution in the data.
- A very good metric to check bias in the imputations.
Fit a linear model for statistical summary
import statsmodels.api as smdiabetes_cc = diabetes.dropna(how='any') X = sm.add_constant(diabetes_cc.iloc[:, :-1]) y = diabetes_cc['Class'] lm = sm.OLS(y, X).fit()
print(lm.summary())
Summary: OLS Regression Results
==============================================================================
Dep. Variable: Class R-squared: 0.346
Model: OLS Adj. R-squared: 0.332
Method: Least Squares F-statistic: 25.30
Date: Wed, 10 Jul 2019 Prob (F-statistic): 2.65e-31
Time: 15:03:19 Log-Likelihood: -177.76
No. Observations: 392 AIC: 373.5
Df Residuals: 383 BIC: 409.3
Df Model: 8
Covariance Type: nonrobust
=====================================================================================
coef std err t P>|t| [0.025 0.975]
<hr />----------------------------------------------------------------------------------
const -1.1027 0.144 -7.681 0.000 -1.385 -0.820
Pregnant 0.0130 0.008 1.549 0.122 -0.003 0.029
Glucose 0.0064 0.001 7.855 0.000 0.005 0.008
Diastolic_BP 5.465e-05 0.002 0.032 0.975 -0.003 0.003
Skin_Fold 0.0017 0.003 0.665 0.506 -0.003 0.007
Serum_Insulin -0.0001 0.000 -0.603 0.547 -0.001 0.000
BMI 0.0093 0.004 2.391 0.017 0.002 0.017
Diabetes_Pedigree 0.1572 0.058 2.708 0.007 0.043 0.271
Age 0.0059 0.003 2.109 0.036 0.000 0.011
R-squared and Coefficients
lm.rsquared_adj
0.33210
lm.params
const -1.102677
Pregnant 0.012953
Glucose 0.006409
Diastolic_BP 0.000055
Skin_Fold 0.001678
Serum_Insulin -0.000123
BMI 0.009325
Diabetes_Pedigree 0.157192
Age 0.005878
dtype: float64
Fit linear model on different imputed DataFrames
# Mean Imputation X = sm.add_constant(diabetes_mean_imputed.iloc[:, :-1]) y = diabetes['Class'] lm_mean = sm.OLS(y, X).fit()# KNN Imputation X = sm.add_constant(diabetes_knn_imputed.iloc[:, :-1]) lm_KNN = sm.OLS(y, X).fit()# MICE Imputation X = sm.add_constant(diabetes_mice_imputed.iloc[:, :-1]) lm_MICE = sm.OLS(y, X).fit()
Comparing R-squared of different imputations
print(pd.DataFrame({'Complete': lm.rsquared_adj,
'Mean Imp.': lm_mean.rsquared_adj,
'KNN Imp.': lm_KNN.rsquared_adj,
'MICE Imp.': lm_MICE.rsquared_adj},
index=['R_squared_adj']))
Complete Mean Imp. KNN Imp. MICE Imp.
R_squared_adj 0.332108 0.313781 0.316543 0.317679
Note: The metrics used here is for linear correlations only. You must use the metrics that are reflective of the data.
Comparing coefficients of different imputations
print(pd.DataFrame({'Complete': lm.params,
'Mean Imp.': lm_mean.params,
'KNN Imp.': lm_KNN.params,
'MICE Imp.': lm_MICE.params}))
Complete Mean Imp. KNN Imp. MICE Imp.
const -1.102677 -1.024005 -1.028035 -1.050023
Pregnant 0.012953 0.020693 0.020047 0.020295
Glucose 0.006409 0.006467 0.006614 0.006871
Diastolic_BP 0.000055 -0.001137 -0.001196 -0.001317
Skin_Fold 0.001678 0.000193 0.001626 0.000807
Serum_Insulin -0.000123 -0.000090 -0.000147 -0.000227
BMI 0.009325 0.014376 0.013239 0.014203
Diabetes_Pedigree 0.157192 0.129282 0.128038 0.129056
Age 0.005878 0.002092 0.002046 0.002097
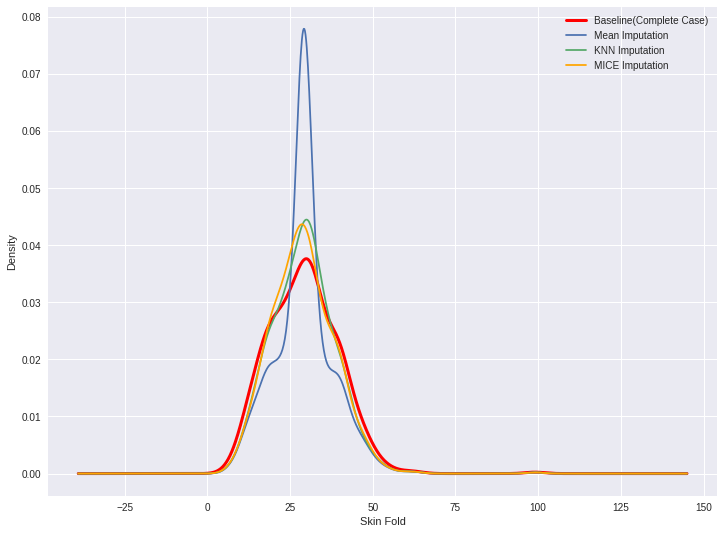
Comparing density plots
diabetes_cc['Skin_Fold'].plot(kind='kde', c='red', linewidth=3)
diabetes_mean_imputed['Skin_Fold'].plot(kind='kde')
diabetes_knn_imputed['Skin_Fold'].plot(kind='kde')
diabetes_mice_imputed['Skin_Fold'].plot(kind='kde')
labels = ['Baseline (Complete Case)', 'Mean Imputation', 'KNN Imputation',
'MICE Imputation']
plt.legend(labels)
plt.xlabel('Skin Fold')
Comparing density plots
Summary
- Applying linear model from the statsmodels package
- Comparing the coefficients and standard errors
- Comparing density plots
Let's practice!
Dealing with Missing Data in Python

