t-SNE for 2-dimensional maps
Unsupervised Learning in Python

Benjamin Wilson
Director of Research at lateral.io
t-SNE for 2-dimensional maps
- t-SNE = "t-distributed stochastic neighbor embedding"
- Maps samples to 2D space (or 3D)
- Map approximately preserves nearness of samples
- Great for inspecting datasets
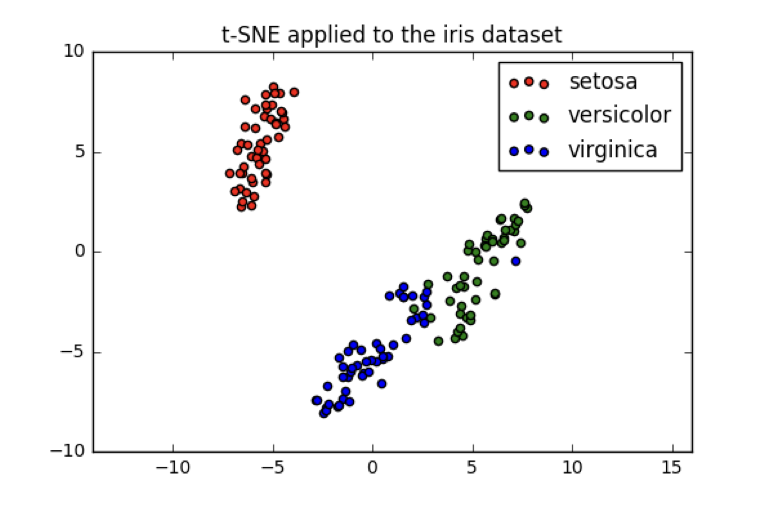
t-SNE on the iris dataset
- Iris dataset has 4 measurements, so samples are 4-dimensional
- t-SNE maps samples to 2D space
- t-SNE didn't know that there were different species
- ... yet kept the species mostly separate
Interpreting t-SNE scatter plots
- "versicolor" and "virginica" harder to distinguish from one another
- Consistent with k-means inertia plot: could argue for 2 clusters, or for 3

t-SNE in sklearn
- 2D NumPy array
samples
print(samples)
[[ 5. 3.3 1.4 0.2]
[ 5. 3.5 1.3 0.3]
[ 4.9 2.4 3.3 1. ]
[ 6.3 2.8 5.1 1.5]
...
[ 4.9 3.1 1.5 0.1]]
- List
speciesgiving species of labels as number (0, 1, or 2)
print(species)
[0, 0, 1, 2, ..., 0]
t-SNE in sklearn
import matplotlib.pyplot as plt from sklearn.manifold import TSNE model = TSNE(learning_rate=100)transformed = model.fit_transform(samples) xs = transformed[:,0] ys = transformed[:,1] plt.scatter(xs, ys, c=species) plt.show()
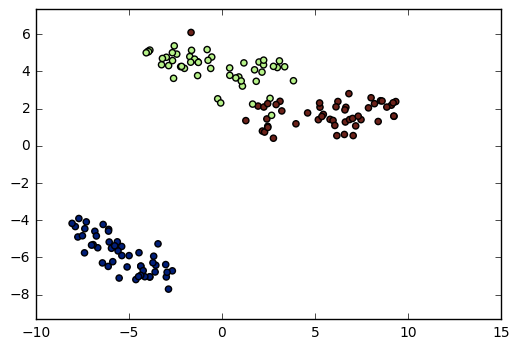
t-SNE has only fit_transform()
- Has a
fit_transform()method - Simultaneously fits the model and transforms the data
- Has no separate
fit()ortransform()methods - Can't extend the map to include new data samples
- Must start over each time!
t-SNE learning rate
- Choose learning rate for the dataset
- Wrong choice: points bunch together
- Try values between 50 and 200
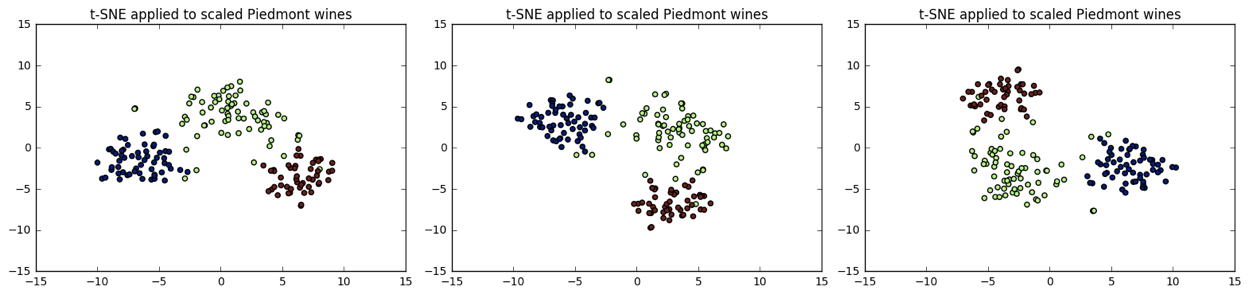
Different every time
- t-SNE features are different every time
- Piedmont wines, 3 runs, 3 different scatter plots!
- ... however: The wine varieties (=colors) have same position relative to one another
Let's practice!
Unsupervised Learning in Python

