Multiple imputation by bootstrapping
Handling Missing Data with Imputations in R

Michal Oleszak
Machine Learning Engineer
Uncertainty from imputation
- Imputation is typically a first step before analysis or modeling.
- Missing values are estimated with some uncertainty.
- This uncertainty should be accounted for in any analyses carried out on imputed data.
In almost half of the studies, key results disappear
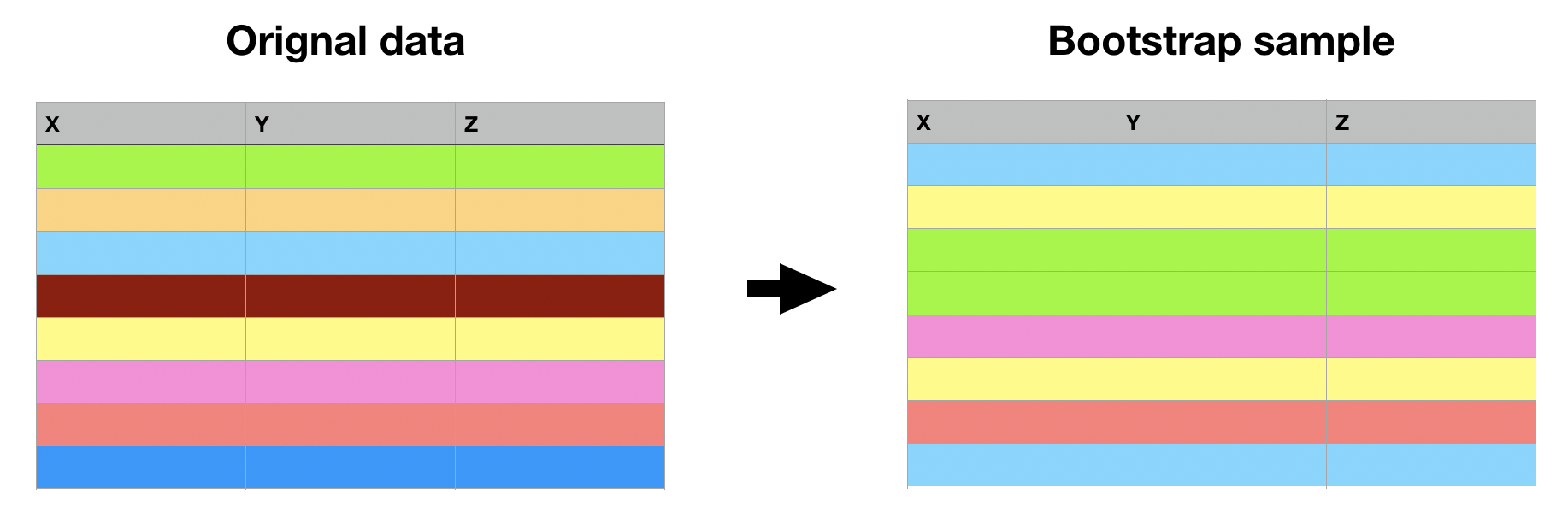
Bootstrap
Bootstrapping = sampling rows with replacement to get original-size data
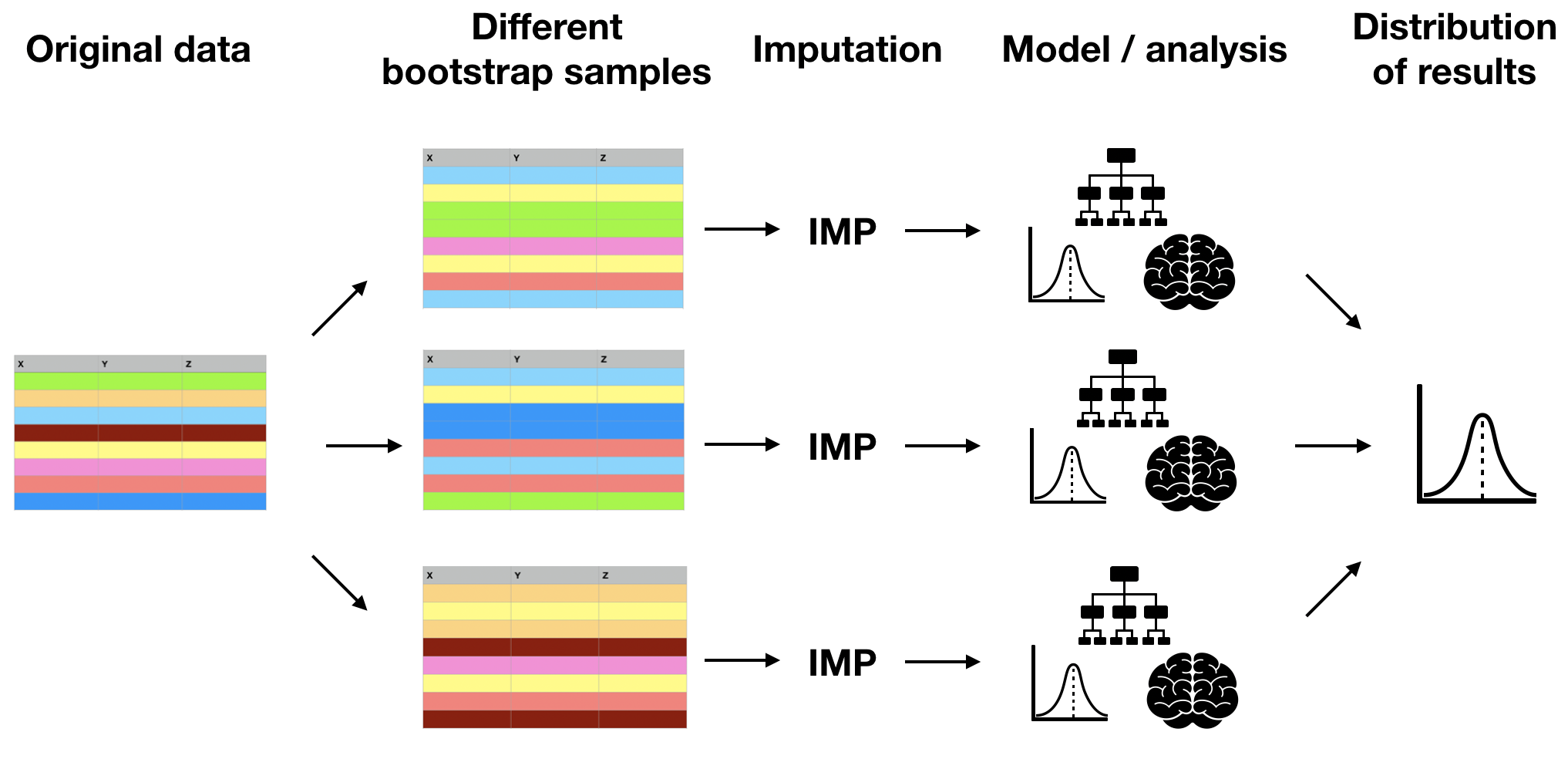
Multiple imputation by bootstrapping
Bootstrapped imputation: pros & cons
Pros:
- Works with any imputation method.
- Can approximate quantities that are hard to compute analytically.
- Work with MCAR and MAR data.
Cons:
- Slow for many replicates or time-consuming computations.
Bootstrapping in practice
calc_correlation <- function(data, indices) {
# Return the correlation coefficient
return(corr_coeff)
}
Bootstrapping in practice
calc_correlation <- function(data, indices) {
# Get bootstrap sample
data_boot <- data[indices, ]
# Return the correlation coefficient
return(corr_coeff)
}
Bootstrapping in practice
calc_correlation <- function(data, indices) {
# Get bootstrap sample
data_boot <- data[indices, ]
# Impute with kNN imputation
data_imp <- kNN(data_boot)
# Return the correlation coefficient
return(corr_coeff)
}
Bootstrapping in practice
calc_correlation <- function(data, indices) {
# Get bootstrap sample
data_boot <- data[indices, ]
# Impute with kNN imputation
data_imp <- kNN(data_boot)
# Calculate correlation between Weight and TotChol
corr_coeff <- cor(data_imp$Weight, data_imp$TotChol)
# Return the correlation coefficient
return(corr_coeff)
}
Bootstrapping in practice
library(boot)
boot_results <- boot(nhanes, statistic = calc_correlation, R = 50)
print(boot_results)
ORDINARY NONPARAMETRIC BOOTSTRAP
Call:
boot(data = nhanes, statistic = calc_correlation, R = 50)
Bootstrap Statistics :
original bias std. error
t1* 0.03028306 0.007385452 0.04207152
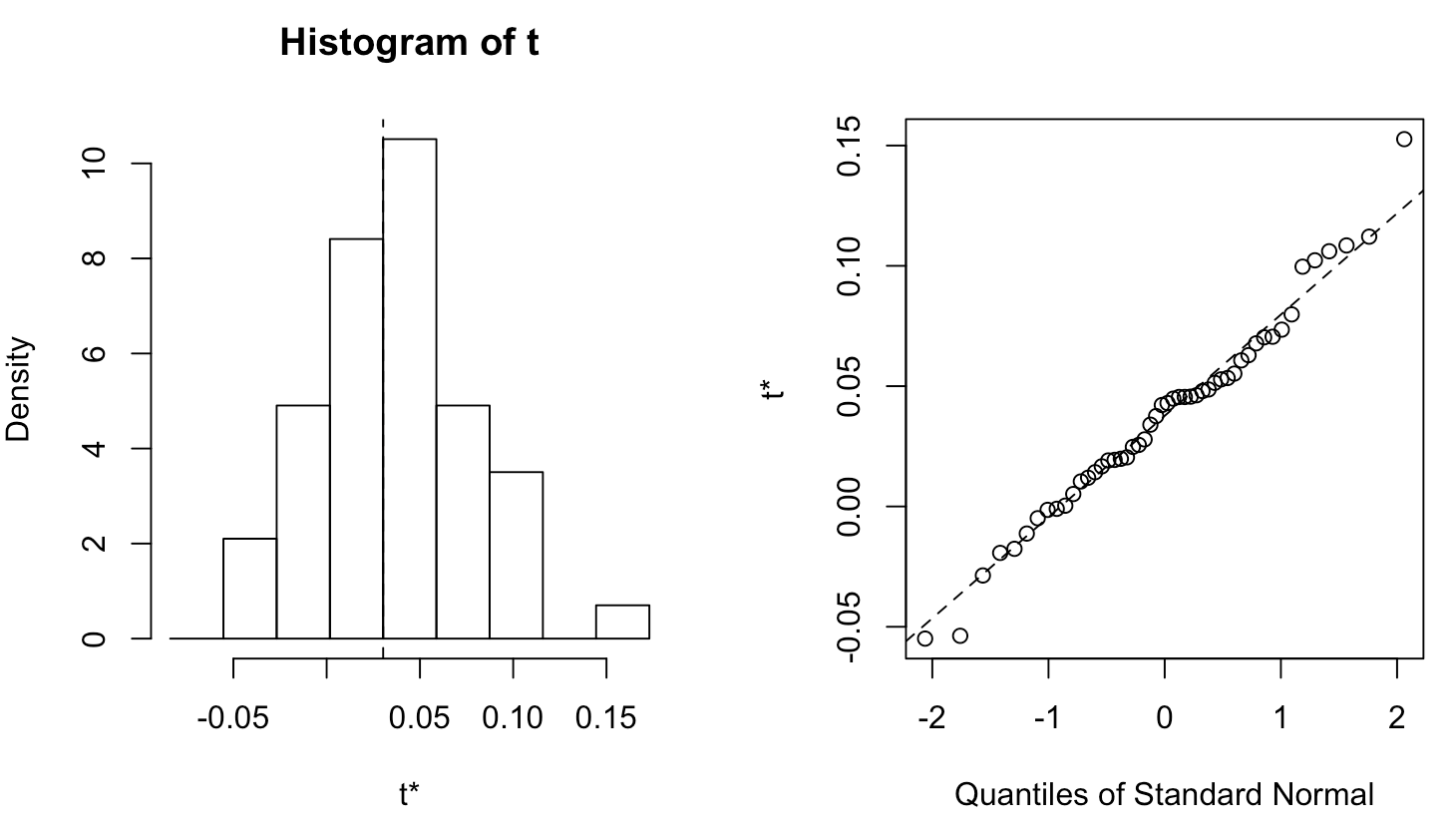
Plotting bootstrap results
plot(boot_results)
Bootstrapping confidence intervals
boot_ci <- boot.ci(boot_results, conf = 0.95, type = "norm")
print(boot_ci)
BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS
Based on 50 bootstrap replicates
CALL :
boot.ci(boot.out = boot_results, conf = 0.95, type = "norm")
Intervals :
Level Normal
95% (-0.0596, 0.1054 )
Calculations and Intervals on Original Scale
Let's practice bootstrapping!
Handling Missing Data with Imputations in R

