Tree-based imputation
Handling Missing Data with Imputations in R

Michal Oleszak
Machine Learning Engineer
Tree-based imputation approach
Use machine learning models to predict missing values!
- Non-parametric approach: no assumptions on relationships between variables.
- Can pick up complex non-linear patterns.
- Often better predictive performance compared to simple statistical models.
This course: missForest package, based on randomForest
Decision trees
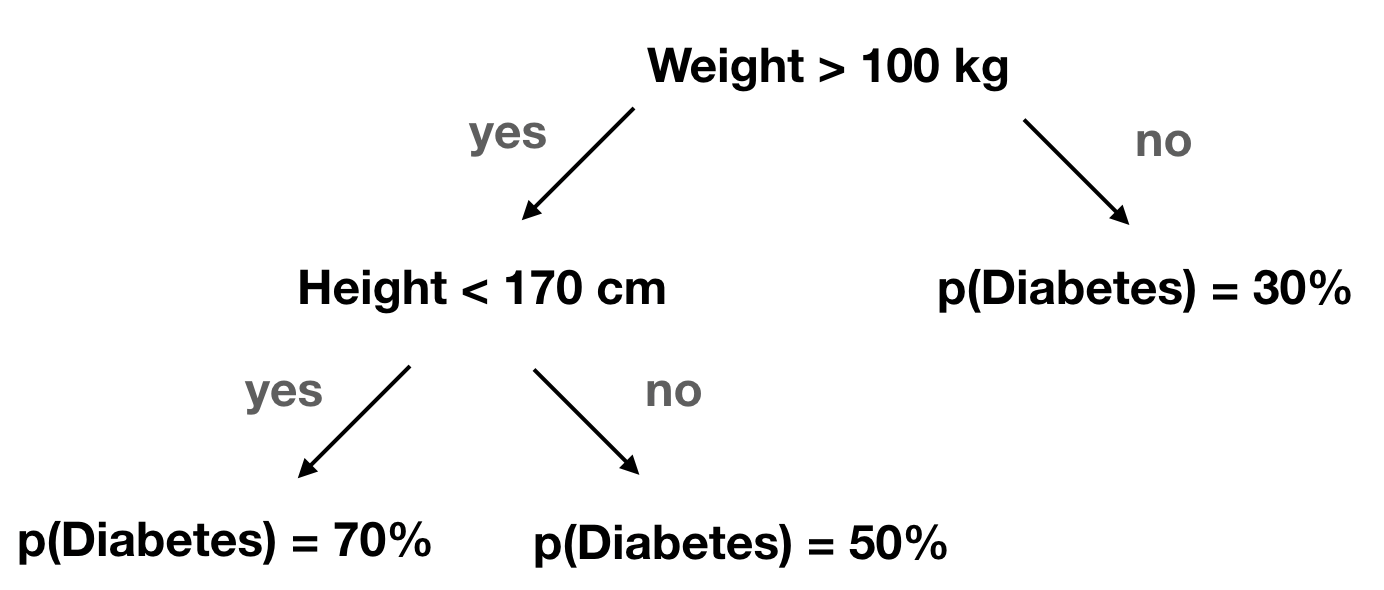
Random forests
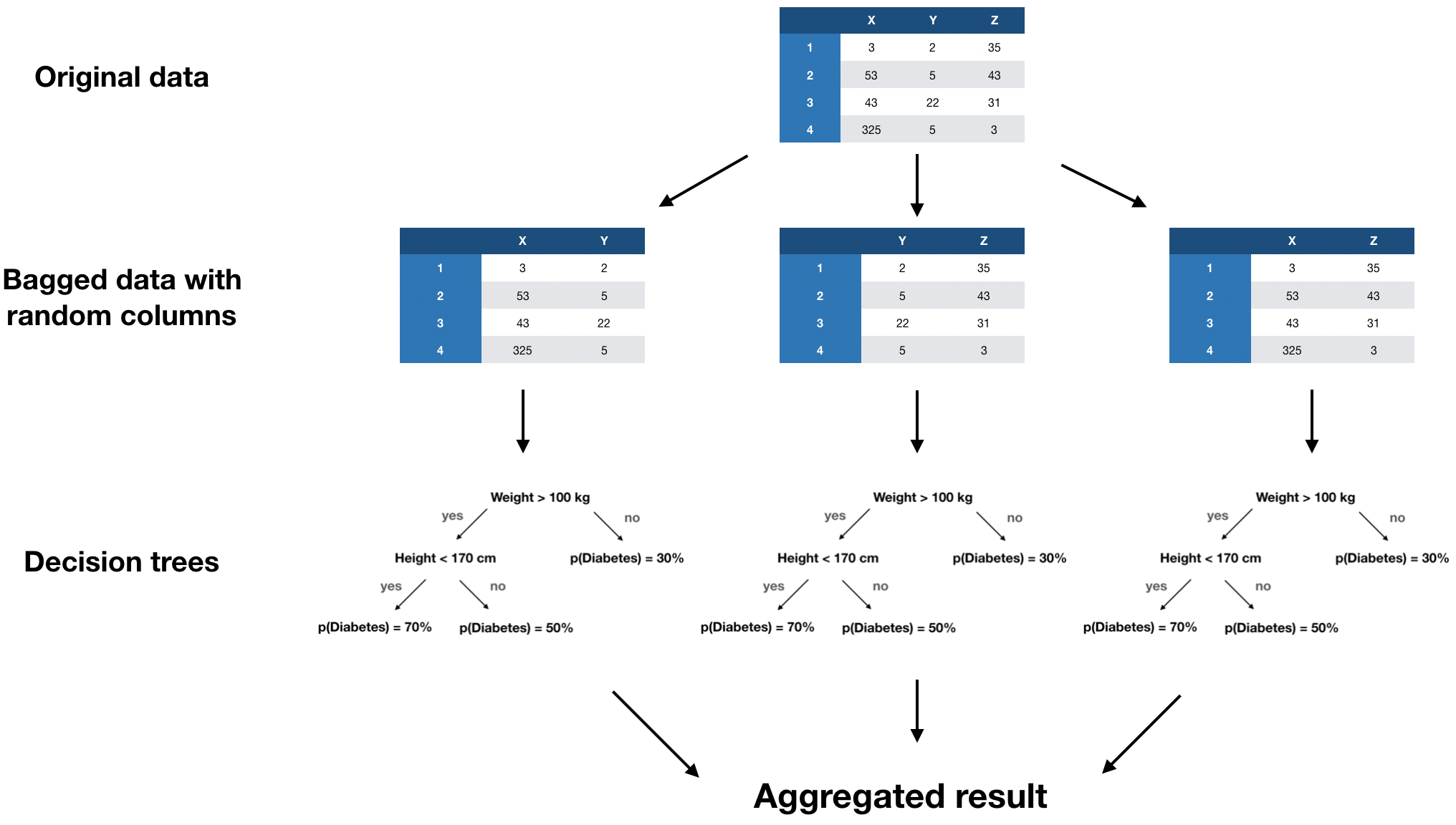
missForest algorithm
- Make an initial guess for missing values with mean imputation.
- Sort the variables in ascending order by the amount of missing values.
- For each variable x:
- Fit a random forest to the observed part of x (using other variables as predictors).
- Use it to predict the missing part of x.
- Repeat step 3. until the imputed values do not change much anymore.
missForest in practice
nhanes %>% is.na() %>% colSums()
Age Gender Weight Height Diabetes TotChol Pulse PhysActive
0 0 9 8 1 85 32 26
library(missForest)
imp_res <- missForest(nhanes)
nhanes_imp <- imp_res$ximp
nhanes_imp %>% is.na() %>% colSums()
Age Gender Weight Height Diabetes TotChol Pulse PhysActive
0 0 0 0 0 0 0 0
Imputation error
missForest() provides an out-of-bag (OOB) imputation error estimate:
- Normalized root mean squared error (NRMSE) for continuous variables.
- Proportion of falsely classified entries (PFC) for categorical variables.
In both cases, good performance leads to a value close to 0 and values around 1 indicate a poor result.
imp_res <- missForest(nhanes)
imp_res$OOBerror
NRMSE PFC
0.147687025 0.003676471
Imputation error
missForest() provides an out-of-bag (OOB) imputation error estimate:
- Normalized root mean squared error (NRMSE) for continuous variables.
- Proportion of falsely classified entries (PFC) for categorical variables.
In both cases, good performance leads to a value close to 0 and values around 1 indicate a poor result.
imp_res <- missForest(nhanes, variablewise = TRUE)
imp_res$OOBerror
MSE PFC MSE MSE PFC MSE MSE MSE
0.00000 0.00000 285.79563 40.42142 0.00735 0.53444 129.03609 0.17576
Speed-accuracy trade-off
Growing multiple random forests can be time-consuming.
Idea: sacrifice some accuracy and reduce the forest size to decrease computation time.
- Reduce the number of trees grown in each forest (
ntreeargument). - Reduce the number of variables used for splitting (
mtryargument).
The effect on computation time differs:
- Reducing
ntreehas a linear effect. - Reducing
mtryincreases speed more when there are many variables.
Speed-accuracy trade-off in practice
Default settings:
start_time <- Sys.time()
imp_res <- missForest(nhanes)
end_time <- Sys.time()
print(imp_res$OOBerror)
print(end_time - start_time)
NRMSE PFC
0.147687025 0.003676471
Time difference of 5.496582 secs
Reduced forests:
start_time <- Sys.time()
imp_res <- missForest(nhanes,
ntree = 10,
mtry = 2)
end_time <- Sys.time()
print(imp_res$OOBerror)
print(end_time - start_time)
NRMSE PFC
0.162420139 0.007425743
Time difference of 0.516367 secs
Let's practice!
Handling Missing Data with Imputations in R

