Mean imputation
Handling Missing Data with Imputations in R

Michal Oleszak
Machine Learning Engineer
Imputation vocabulary
Imputation = making an educated guess about what the missing values might be
- Donor-based imputation - missing values are filled in using other, complete observations.
- Model-based imputation - missing values are predicted with a statistical or machine learning model.
This chapter focuses on donor-based methods:
- Mean imputation
- Hot-deck imputation
- kNN imputation
Mean imputation
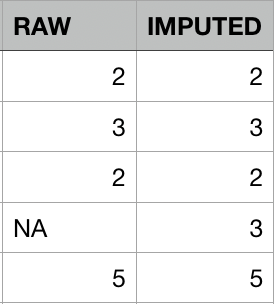
Mean imputation works well for time-series data that randomly fluctuate around a long-term average.
For cross-sectional data, mean imputation is often a very poor choice:
- Destroys relations between variables.
- There is no variance in the imputed values.
Mean imputation in practice
Task: mean-impute Height and Weight in NHANES data.
- Create binary indicators for whether each value was originally missing.
nhanes <- nhanes %>%
mutate(Height_imp = ifelse(is.na(Height), TRUE, FALSE)) %>%
mutate(Weight_imp = ifelse(is.na(Weight), TRUE, FALSE))
- Replace missing values in
HeightandWeightwith their respective means.
nhanes_imp <- nhanes %>%
mutate(Height = ifelse(is.na(Height), mean(Height, na.rm = TRUE), Height)) %>%
mutate(Weight = ifelse(is.na(Weight), mean(Weight, na.rm = TRUE), Weight))
Mean-imputed NHANES data
nhanes_imp %>%
select(Weight, Height, Height_imp, Weight_imp) %>%
head()
Weight Height Height_imp Weight_imp
1 73.20000 166.2499 TRUE FALSE
2 72.30000 166.2499 TRUE FALSE
3 57.70000 158.9000 FALSE FALSE
4 88.90000 183.3000 FALSE FALSE
5 45.10000 157.6000 FALSE FALSE
6 66.77065 158.4000 FALSE TRUE
Assessing imputation quality: margin plot
nhanes_imp %>% select(Weight, Height, Height_imp, Weight_imp) %>% marginplot(delimiter="imp")
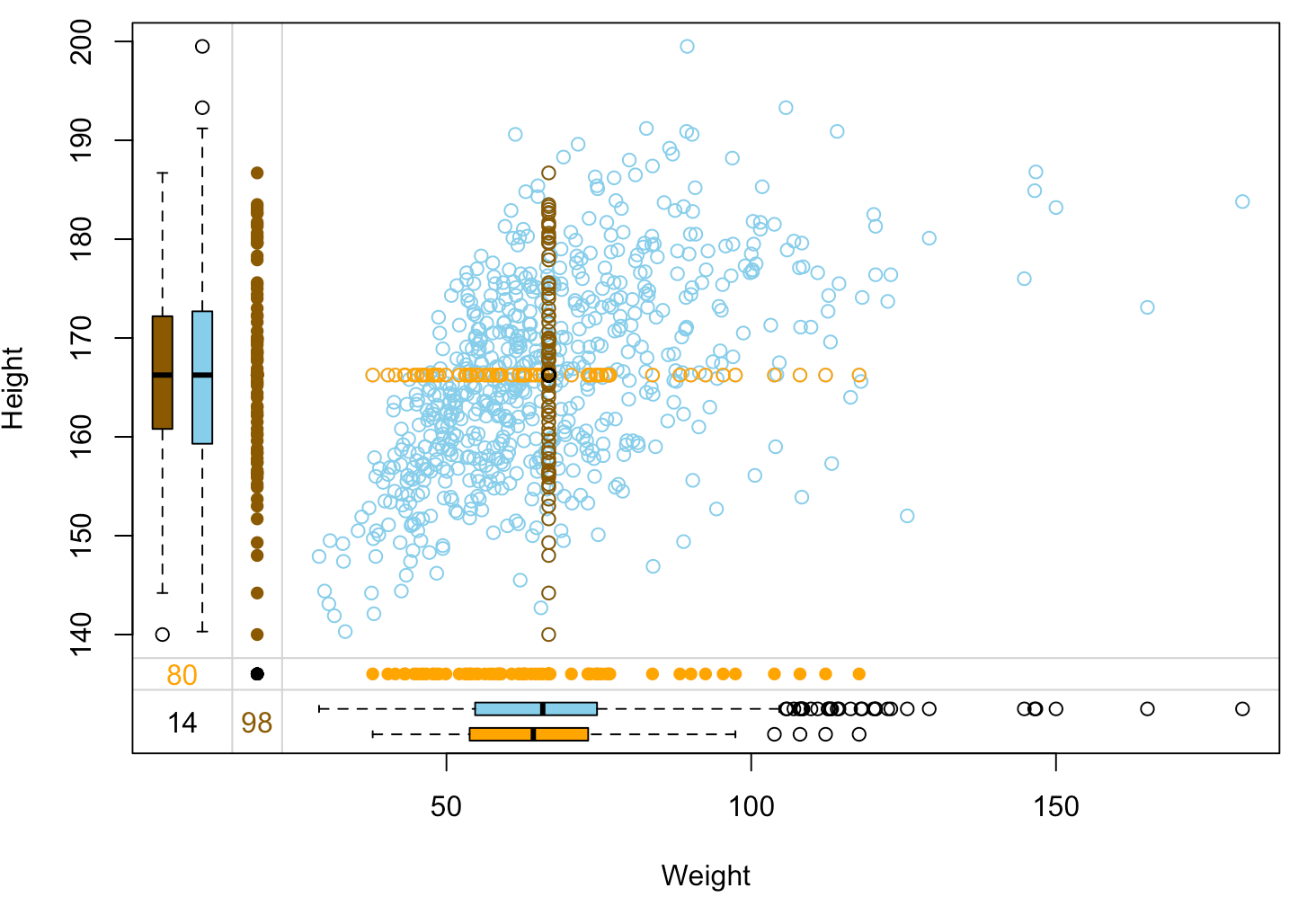
Troubles with mean imputation
Destroying relation between variables:
- After mean-imputing
HeightandWeight, their positive correlation is weaker. - Models predicting one using the other will be fooled by the outlying imputed values and will produce biased results.
No variability in imputed data:
- With less variance in the data, all standard errors will be underestimated. This prevents reliable hypothesis testing and calculating confidence intervals.
Median and mode imputation
- Instead of the mean, one might impute with a median or a mode.
- Median imputation is a better choice when there are outliers in the data.
- For categorical variables, we cannot compute neither mean or median, so we use the mode instead.
- Both median and mode imputation present the same drawbacks as mean imputation.
Let's practice!
Handling Missing Data with Imputations in R

