Multiple imputation by chained equations
Handling Missing Data with Imputations in R

Michal Oleszak
Machine Learning Engineer
The MICE algorithm
1 van Buuren, S., & Groothuis-Oudshoorn, C. G. M. (2011). mice: Multivariate Imputation by Chained Equations in R. Journal of statistical software, 45(3).
MICE: pros & cons
Pros:
- Requires fewer replications than the bootstrap.
- Works for MAR and MCAR data.
Allows for sensitivity analysis towards MNAR data.
Cons:
- Only works with selected imputation methods.
- Requires more tuning effort (model selection, choosing predictors).
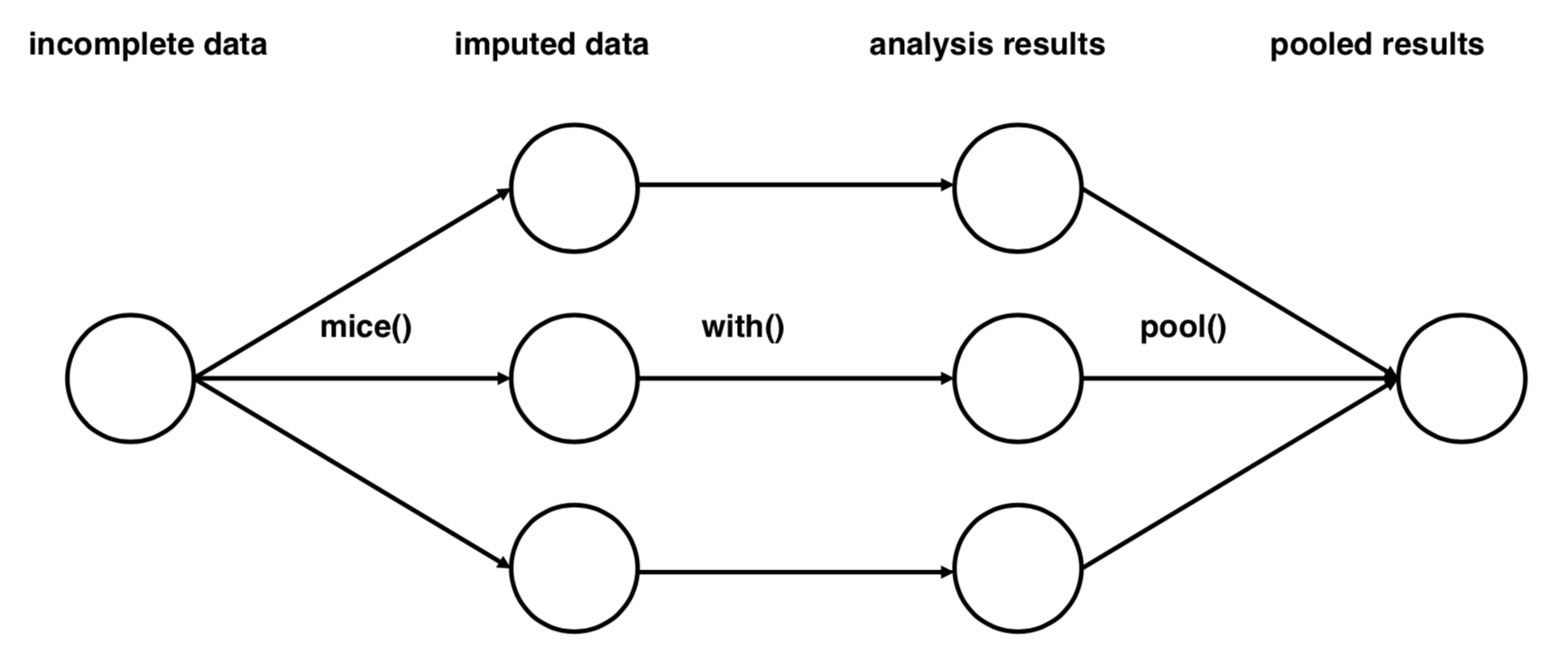
The mice flow: mice - with - pool
Impute nhanes 20 times:
library(mice)
nhanes_multiimp <- mice(nhanes, m = 20)
Fit a linear regression model to each imputed data set:
lm_multiimp <- with(nhanes_multiimp, lm(Weight ~ Height + TotChol + PhysActive))
Pool regression results:
lm_pooled <- pool(lm_multiimp)
Analyzing pooled results
summary(lm_pooled, conf.int = TRUE, conf.level = 0.95)
estimate std.error statistic df p.value 2.5 % 97.5 %
(Intercept) -122.964 10.933 -11.247 735.389 0.000 -144.428 -101.500
Height 1.086 0.060 18.158 796.106 0.000 0.968 1.203
TotChol 2.653 0.884 3.003 305.460 0.003 0.915 4.392
PhysActive -1.746 1.422 -1.228 733.957 0.220 -4.536 1.045
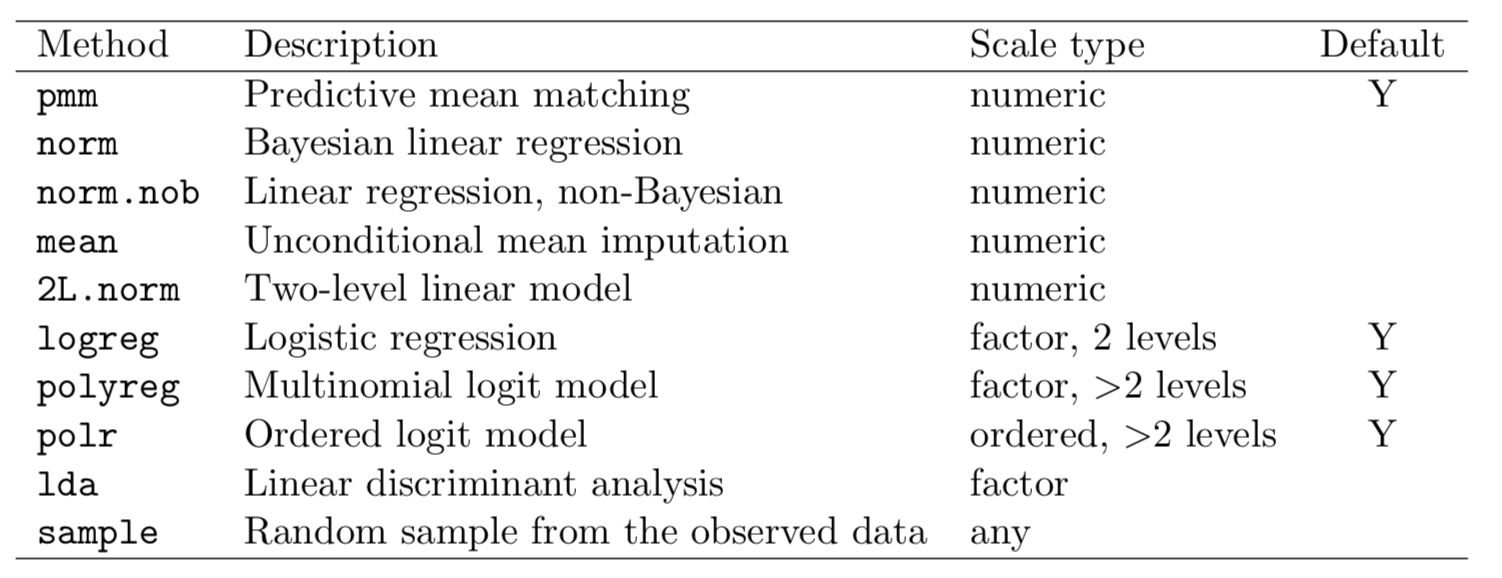
MICE: available methods
1 van Buuren, S., & Groothuis-Oudshoorn, C. G. M. (2011). mice: Multivariate Imputation by Chained Equations in R. Journal of statistical software, 45(3).
Choosing methods per variable type
mice() takes an argument defaultMethod: a vector of 4 strings, specifying methods for:
- Continuous variables
- Binary variables
- Categorical variables (unordered factors)
- Factor variables (ordered factors)
nhanes_multiimp <- mice(nhanes, m = 20,
defaultMethod = c("pmm", "logreg", "polyreg", "polr"))
Predictor matrix
The predictorMatrix governs which variables are used to impute other variables.
nhanes_multiimp <- mice(nhanes, m = 20)
nhanes_multiimp$predictorMatrix
Age Gender Weight Height Diabetes TotChol Pulse PhysActive
Age 0 1 1 1 1 1 1 1
Gender 1 0 1 1 1 1 1 1
Weight 1 1 0 1 1 1 1 1
Height 1 1 1 0 1 1 1 1
Diabetes 1 1 1 1 0 1 1 1
TotChol 1 1 1 1 1 0 1 1
Pulse 1 1 1 1 1 1 0 1
PhysActive 1 1 1 1 1 1 1 0
Choosing predictors for each variable
- Ideally, a proper model selection should be performed.
- A quick alternative: use variables correlated with the target.
pred_mat <- quickpred(nhanes, mincor = 0.25)
nhanes_multiimp <- mice(nhanes, m = 20, predictorMatrix = pred_mat)
print(pred_mat)
Age Gender Weight Height Diabetes TotChol Pulse PhysActive
Age 0 0 0 0 0 0 0 0
Gender 0 0 0 0 0 0 0 0
Weight 1 1 0 0 0 0 1 0
...
Let's practice imputing with MICE!
Handling Missing Data with Imputations in R

