Replicating data variability
Handling Missing Data with Imputations in R

Michal Oleszak
Machine Learning Engineer
Variability in imputed data
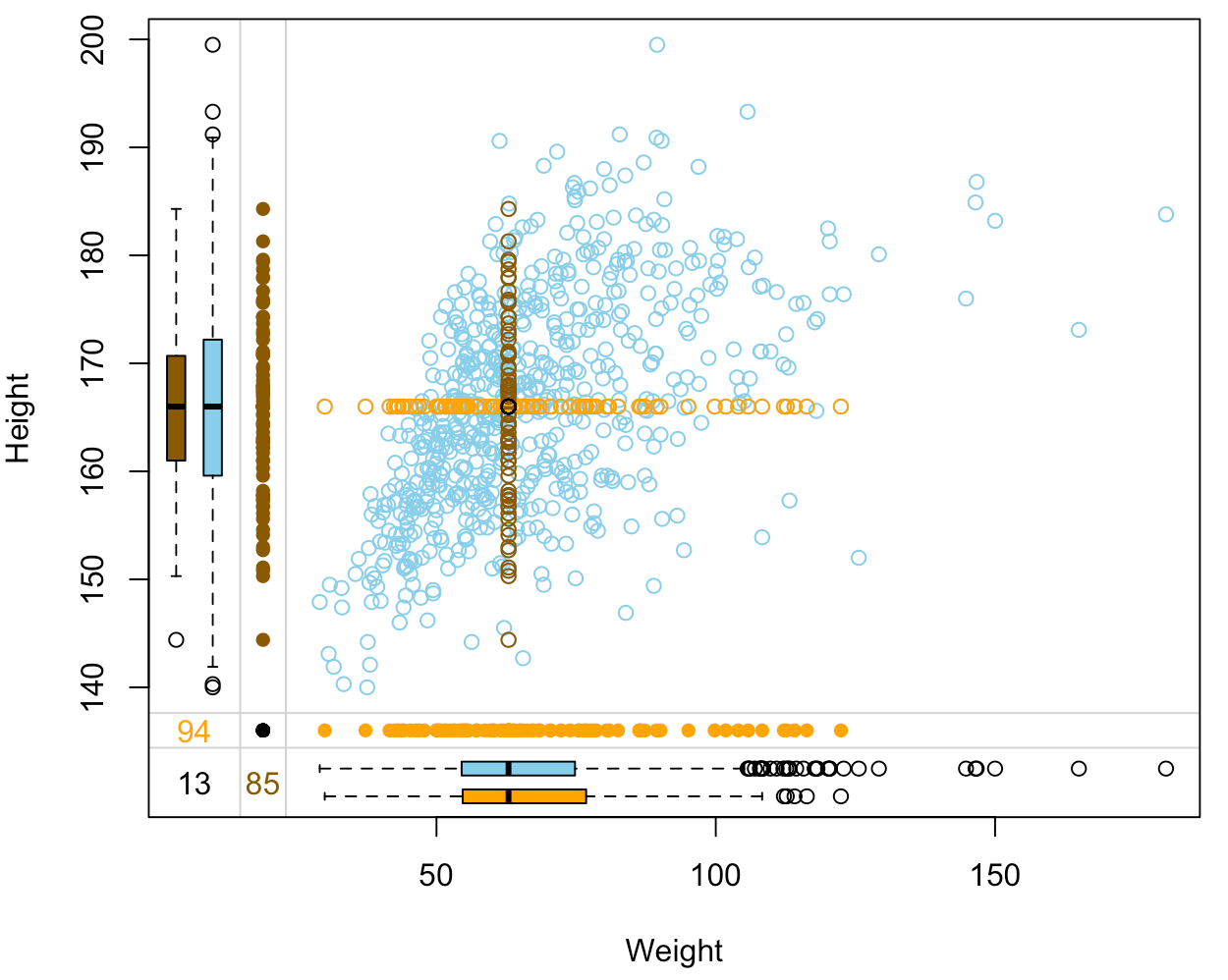
- No variability in imputed data.
- We would like the imputation to replicate the variability of observed data.
- In model-based imputation, the same values of predictors result in the same imputed value.
- Solution: drawing from conditional distributions.
What is a prediction
Most statistical models estimate the conditional distribution of the response variable:
$p(y|X)$
To make a single prediction, the conditional distribution is summarized:
- Linear regression: expected value of the conditional distribution.
- Logistic regression: class with the highest probability.
Instead, we can draw from these distributions to increase variability.
Drawing from conditional distributions
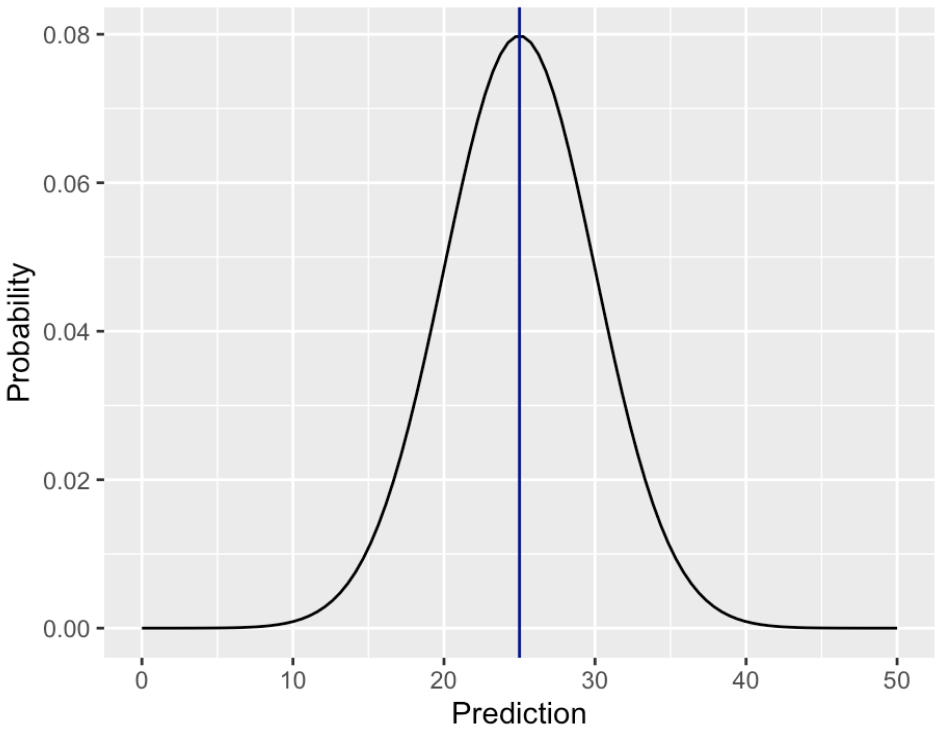
Drawing from conditional distributions
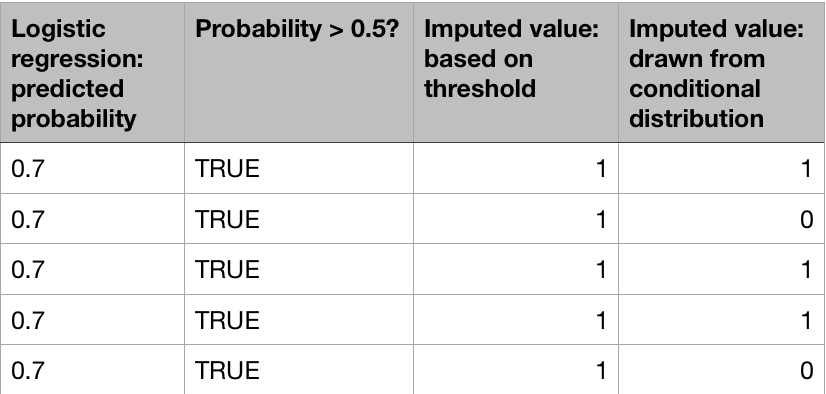
Logistic regression imputation
Task: impute PhysActive from nhanes data with logistic regression.
nhanes_imp <- hotdeck(nhanes)
missing_physactive <- is.na(nhanes$PhysActive)
Logistic regression imputation
Task: impute PhysActive from nhanes data with logistic regression.
nhanes_imp <- hotdeck(nhanes)
missing_physactive <- is.na(nhanes$PhysActive)
logreg_model <- glm(PhysActive ~ Age + Weight + Pulse,
data = nhanes_imp, family = binomial)
Logistic regression imputation
Task: impute PhysActive from nhanes data with logistic regression.
nhanes_imp <- hotdeck(nhanes)
missing_physactive <- is.na(nhanes$PhysActive)
logreg_model <- glm(PhysActive ~ Age + Weight + Pulse,
data = nhanes_imp, family = binomial)
preds <- predict(logreg_model, type = "response")
Logistic regression imputation
Task: impute PhysActive from nhanes data with logistic regression.
nhanes_imp <- hotdeck(nhanes)
missing_physactive <- is.na(nhanes$PhysActive)
logreg_model <- glm(PhysActive ~ Age + Weight + Pulse,
data = nhanes_imp, family = binomial)
preds <- predict(logreg_model, type = "response")
preds <- ifelse(preds >= 0.5, 1, 0)
Logistic regression imputation
Task: impute PhysActive from nhanes data with logistic regression.
nhanes_imp <- hotdeck(nhanes)
missing_physactive <- is.na(nhanes$PhysActive)
logreg_model <- glm(PhysActive ~ Age + Weight + Pulse,
data = nhanes_imp, family = binomial)
preds <- predict(logreg_model, type = "response")
preds <- ifelse(preds >= 0.5, 1, 0)
nhanes_imp[missing_physactive, "PhysActive"] <- preds[missing_physactive]
Logistic regression imputation
Variability of imputed data:
table(preds[missing_physactive])
1
26
Variability of observed PhysActive data:
table(nhanes$PhysActive)
0 1
181 610
Drawing from class probabilities
nhanes_imp <- hotdeck(nhanes)
missing_physactive <- is.na(nhanes$PhysActive)
logreg_model <- glm(PhysActive ~ Age + Weight + Pulse,
data = nhanes_imp, family = binomial)
preds <- predict(logreg_model, type = "response")
preds <- ifelse(preds >= 0.5, 1, 0)
nhanes_imp[missing_physactive, "PhysActive"] <- preds[missing_physactive]
Drawing from class probabilities
nhanes_imp <- hotdeck(nhanes)
missing_physactive <- is.na(nhanes$PhysActive)
logreg_model <- glm(PhysActive ~ Age + Weight + Pulse,
data = nhanes_imp, family = binomial)
preds <- predict(logreg_model, type = "response")
nhanes_imp[missing_physactive, "PhysActive"] <- preds[missing_physactive]
Drawing from class probabilities
nhanes_imp <- hotdeck(nhanes)
missing_physactive <- is.na(nhanes$PhysActive)
logreg_model <- glm(PhysActive ~ Age + Weight + Pulse,
data = nhanes_imp, family = binomial)
preds <- predict(logreg_model, type = "response")
preds <- rbinom(length(preds), size = 1, prob = preds)
nhanes_imp[missing_physactive, "PhysActive"] <- preds[missing_physactive]
Drawing from class probabilities
Variability of imputed data:
table(preds[missing_physactive])
0 1
5 21
Variability of observed PhysActive data:
table(nhanes$PhysActive)
0 1
181 610
Let's practice replicating data variability!
Handling Missing Data with Imputations in R

