Outliers, leverage, and influence
Introduction to Regression in R

Richie Cotton
Data Evangelist at DataCamp
Roach dataset
roach <- fish %>%
filter(species == "Roach")
| species | length_cm | mass_g |
|---|---|---|
| Roach | 12.9 | 40 |
| Roach | 16.5 | 69 |
| Roach | 17.5 | 78 |
| Roach | 18.2 | 87 |
| Roach | 18.6 | 120 |
| ... | ... | ... |
Which points are outliers?
ggplot(roach, aes(length_cm, mass_g)) +
geom_point() +
geom_smooth(method = "lm", se = FALSE)
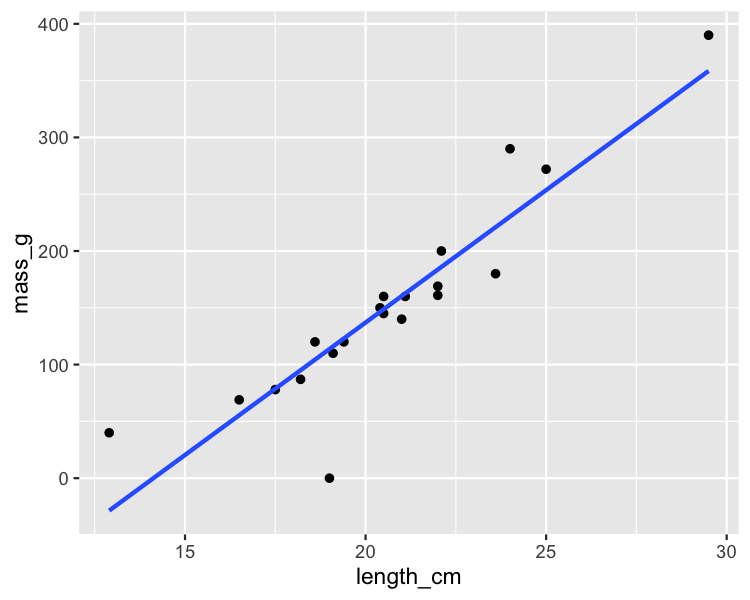
Extreme explanatory values
roach %>%
mutate(
has_extreme_length = length_cm < 15 | length_cm > 26
) %>%
ggplot(aes(length_cm, mass_g)) +
geom_point(aes(color = has_extreme_length)) +
geom_smooth(method = "lm", se = FALSE)
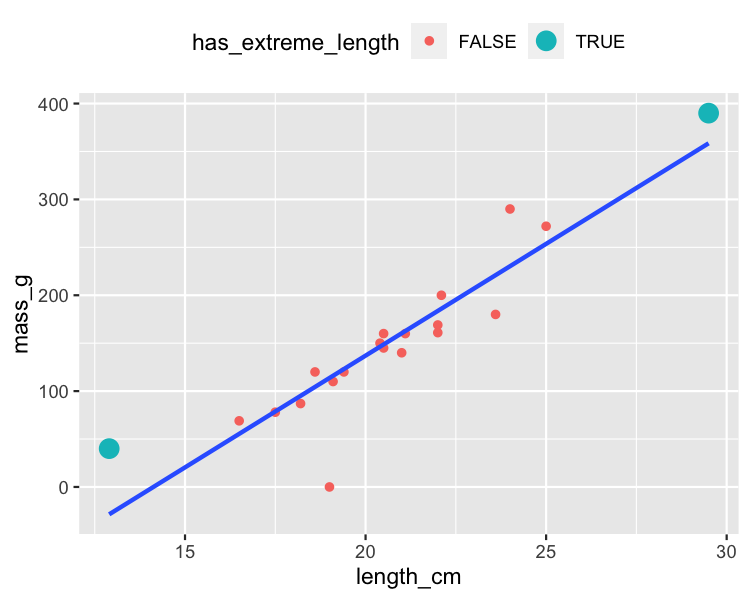
Response values away from the regression line
roach %>%
mutate(
has_extreme_length = length_cm < 15 | length_cm > 26,
has_extreme_mass = mass_g < 1
) %>%
ggplot(aes(length_cm, mass_g)) +
geom_point(
aes(
color = has_extreme_length,
shape = has_extreme_mass
)
) +
geom_smooth(method = "lm", se = FALSE)
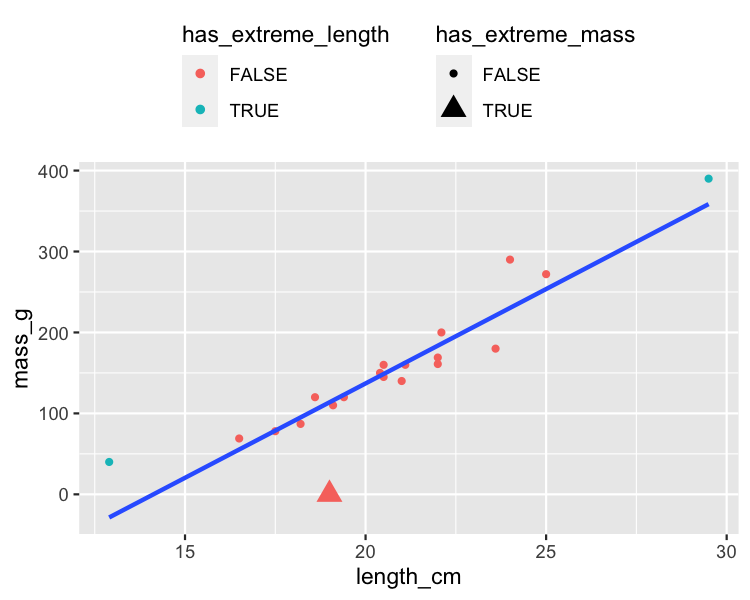
Leverage
Leverage is a measure of how extreme the explanatory variable values are.
mdl_roach <- lm(mass_g ~ length_cm, data = roach)
hatvalues(mdl_roach)
1 2 3 4 5 6 7
0.3137 0.1255 0.0935 0.0763 0.0684 0.0619 0.0605
8 9 10 11 12 13 14
0.0568 0.0503 0.0501 0.0501 0.0506 0.0509 0.0581
15 16 17 18 19 20
0.0581 0.0593 0.0884 0.0995 0.1334 0.3947
The .hat column
library(broom)
augment(mdl_roach)
# A tibble: 20 × 8
mass_g length_cm .fitted .resid .hat .sigma .cooksd .std.resid
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 40 12.9 -28.6 68.6 0.314 33.8 1.07 2.17
2 69 16.5 55.4 13.6 0.126 39.1 0.0104 0.381
3 78 17.5 78.7 -0.711 0.0935 39.3 0.0000197 -0.0196
4 87 18.2 95.0 -8.03 0.0763 39.2 0.00198 -0.219
5 120 18.6 104. 15.6 0.0684 39.1 0.00661 0.424
...
Highly leveraged roaches
mdl_roach %>%
augment() %>%
select(mass_g, length_cm, leverage = .hat) %>%
arrange(desc(leverage)) %>%
head()
# A tibble: 6 x 3
mass_g length_cm leverage
<dbl> <dbl> <dbl>
1 390 29.5 0.395 # really long roach
2 40 12.9 0.314 # really short roach
3 272 25 0.133
4 69 16.5 0.126
5 290 24 0.0995
6 78 17.5 0.0935
Influence
Influence measures how much the model would change if you left the observation out of the dataset when modeling.

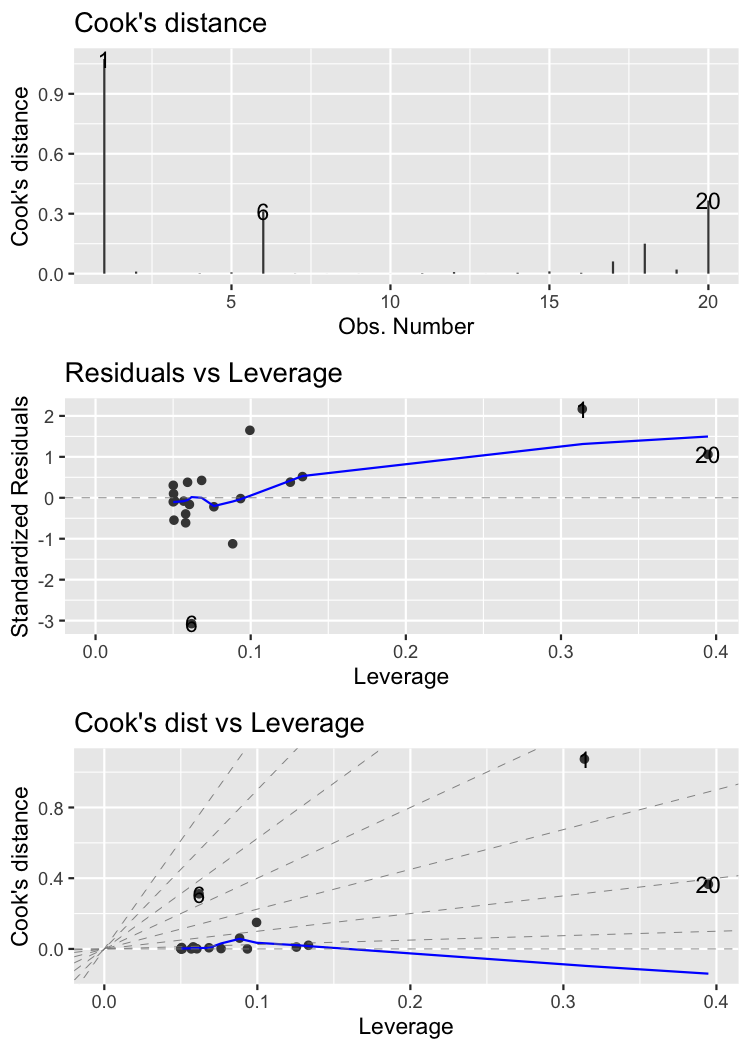
Cook's distance
Cook's distance is the most common measure of influence.
cooks.distance(mdl_roach)
1 2 3 4 5 6
1.07e+00 1.04e-02 1.97e-05 1.98e-03 6.61e-03 3.12e-01
7 8 9 10 11 12
8.53e-04 1.99e-04 2.57e-04 2.56e-04 2.45e-03 7.95e-03
13 14 15 16 17 18
1.37e-04 4.82e-03 1.15e-02 4.52e-03 6.12e-02 1.50e-01
19 20
2.06e-02 3.66e-01
The .cooksd column
library(broom)
augment(mdl_roach)
# A tibble: 20 x 9
mass_g length_cm .fitted .se.fit .resid .hat .sigma .cooksd .std.resid
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 40 12.9 -28.6 21.4 68.6 0.314 33.8 1.07 2.17
2 69 16.5 55.4 13.5 13.6 0.126 39.1 0.0104 0.381
3 78 17.5 78.7 11.7 -0.711 0.0935 39.3 0.0000197 -0.0196
4 87 18.2 95.0 10.5 -8.03 0.0763 39.2 0.00198 -0.219
5 120 18.6 104. 9.98 15.6 0.0684 39.1 0.00661 0.424
...
Most influential roaches
mdl_roach %>%
augment() %>%
select(mass_g, length_cm, cooks_dist = .cooksd) %>%
arrange(desc(cooks_dist)) %>%
head()
# A tibble: 6 x 3
mass_g length_cm cooks_dist
<dbl> <dbl> <dbl>
1 40 12.9 1.07 # really short roach
2 390 29.5 0.366 # really long roach
3 0 19 0.312 # zero mass roach
4 290 24 0.150
5 180 23.6 0.0612
6 272 25 0.0206
Removing the most influential roach
roach_not_short <- roach %>%
filter(length != 12.9)
ggplot(roach, aes(length_cm, mass_g)) +
geom_point() +
geom_smooth(method = "lm", se = FALSE) +
geom_smooth(
method = "lm", se = FALSE,
data = roach_not_short, color = "red"
)
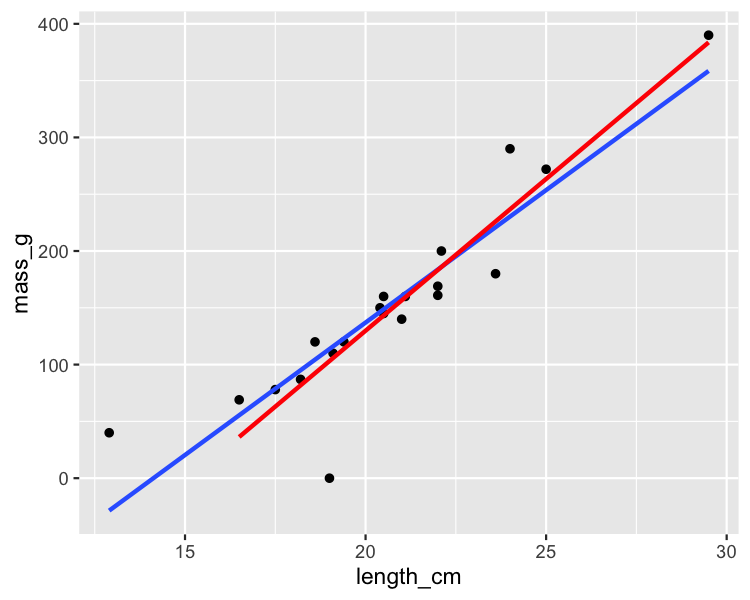
autoplot()
autoplot(
mdl_roach,
which = 4:6,
nrow = 3,
ncol = 1
)
Let's practice!
Introduction to Regression in R

