Models for each category
Intermediate Regression with statsmodels in Python

Maarten Van den Broeck
Content Developer at DataCamp
Four categories
print(fish["species"].unique())
array(['Bream', 'Roach', 'Perch', 'Pike'], dtype=object)
Splitting the dataset
bream = fish[fish["species"] == "Bream"]
perch = fish[fish["species"] == "Perch"]
pike = fish[fish["species"] == "Pike"]
roach = fish[fish["species"] == "Roach"]
Four models
mdl_bream = ols("mass_g ~ length_cm", data=bream).fit()
print(mdl_bream.params)
Intercept -1035.3476
length_cm 54.5500
mdl_perch = ols("mass_g ~ length_cm", data=perch).fit()
print(mdl_perch.params)
Intercept -619.1751
length_cm 38.9115
mdl_pike = ols("mass_g ~ length_cm", data=pike).fit()
print(mdl_pike.params)
Intercept -1540.8243
length_cm 53.1949
mdl_roach = ols("mass_g ~ length_cm", data=roach).fit()
print(mdl_roach.params)
Intercept -329.3762
length_cm 23.3193
Explanatory data
explanatory_data = pd.DataFrame(
{"length_cm": np.arange(5, 61, 5)})
print(explanatory_data)
length_cm
0 5
1 10
2 15
3 20
4 25
5 30
6 35
7 40
8 45
9 50
10 55
11 60
Making predictions
prediction_data_bream = explanatory_data.assign(mass_g = mdl_bream.predict(explanatory_data),species = "Bream")
prediction_data_perch = explanatory_data.assign(mass_g = mdl_perch.predict(explanatory_data),species = "Perch")
prediction_data_pike = explanatory_data.assign(mass_g = mdl_pike.predict(explanatory_data),species = "Pike")
prediction_data_roach = explanatory_data.assign(mass_g = mdl_roach.predict(explanatory_data),species = "Roach")
Concatenating predictions
prediction_data = pd.concat([prediction_data_bream,
prediction_data_roach,
prediction_data_perch,
prediction_data_pike])
length_cm mass_g species
0 5 -762.597660 Bream
1 10 -489.847756 Bream
2 15 -217.097851 Bream
3 20 55.652054 Bream
4 25 328.401958 Bream
5 30 601.151863 Bream
...
3 20 -476.926955 Pike
4 25 -210.952626 Pike
5 30 55.021703 Pike
6 35 320.996032 Pike
7 40 586.970362 Pike
8 45 852.944691 Pike
9 50 1118.919020 Pike
10 55 1384.893349 Pike
11 60 1650.867679 Pike
Visualizing predictions
sns.lmplot(x="length_cm", y="mass_g", data=fish, hue="species", ci=None)plt.show()
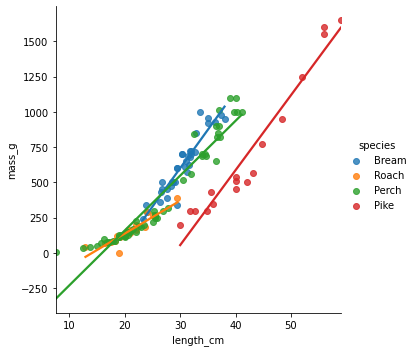
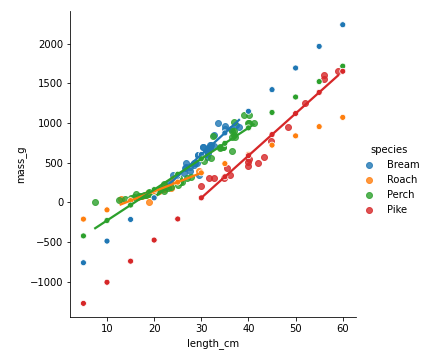
Adding in your predictions
sns.lmplot(x="length_cm",
y="mass_g",
data=fish,
hue="species",
ci=None)
sns.scatterplot(x="length_cm",
y="mass_g",
data=prediction_data,
hue="species",
ci=None,
legend=False)
plt.show()
Coefficient of determination
mdl_fish = ols("mass_g ~ length_cm + species",
data=fish).fit()
print(mdl_fish.rsquared_adj)
0.917
print(mdl_bream.rsquared_adj)
0.874
print(mdl_perch.rsquared_adj)
0.917
print(mdl_pike.rsquared_adj)
0.941
print(mdl_roach.rsquared_adj)
0.815
Residual standard error
print(np.sqrt(mdl_fish.mse_resid))
103
print(np.sqrt(mdl_bream.mse_resid))
74.2
print(np.sqrt(mdl_perch.mse_resid))
100
print(np.sqrt(mdl_pike.mse_resid))
120
print(np.sqrt(mdl_roach.mse_resid))
38.2
Let's practice!
Intermediate Regression with statsmodels in Python

