Cross-validation
Machine Learning with Tree-Based Models in R

Sandro Raabe
Data Scientist
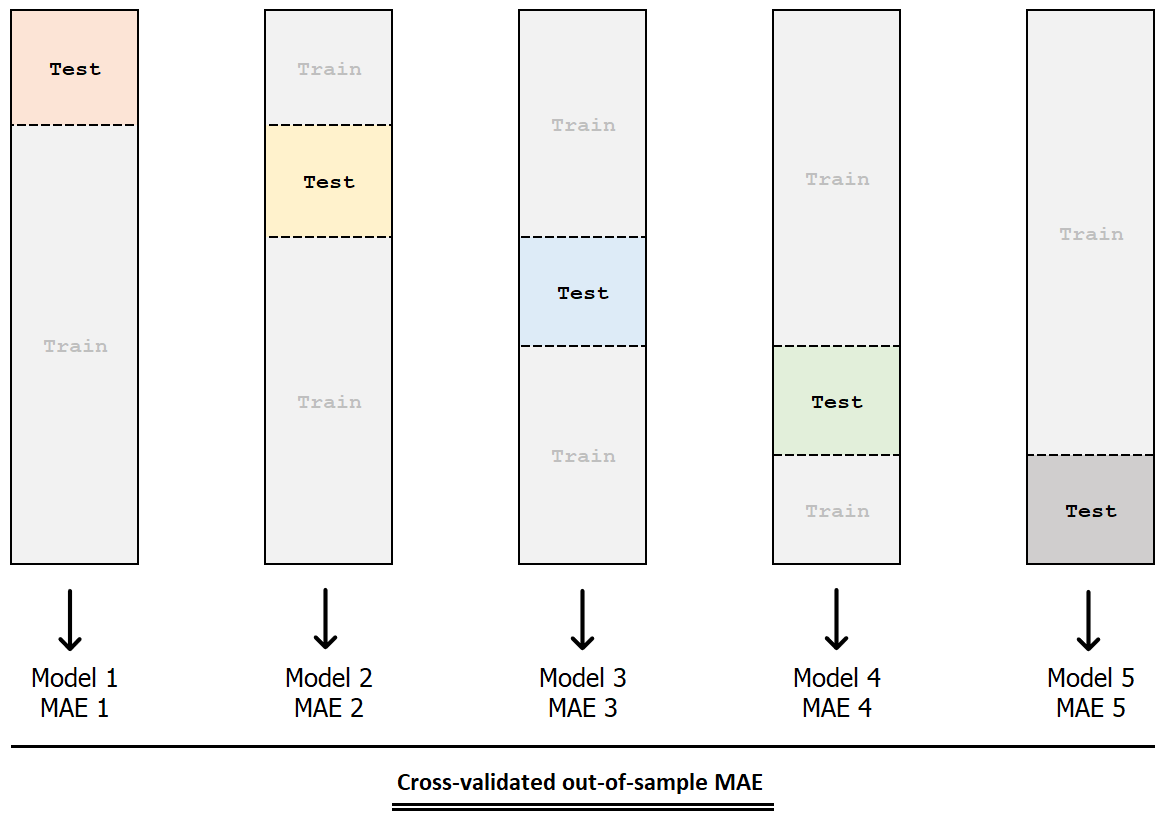
k-fold cross-validation
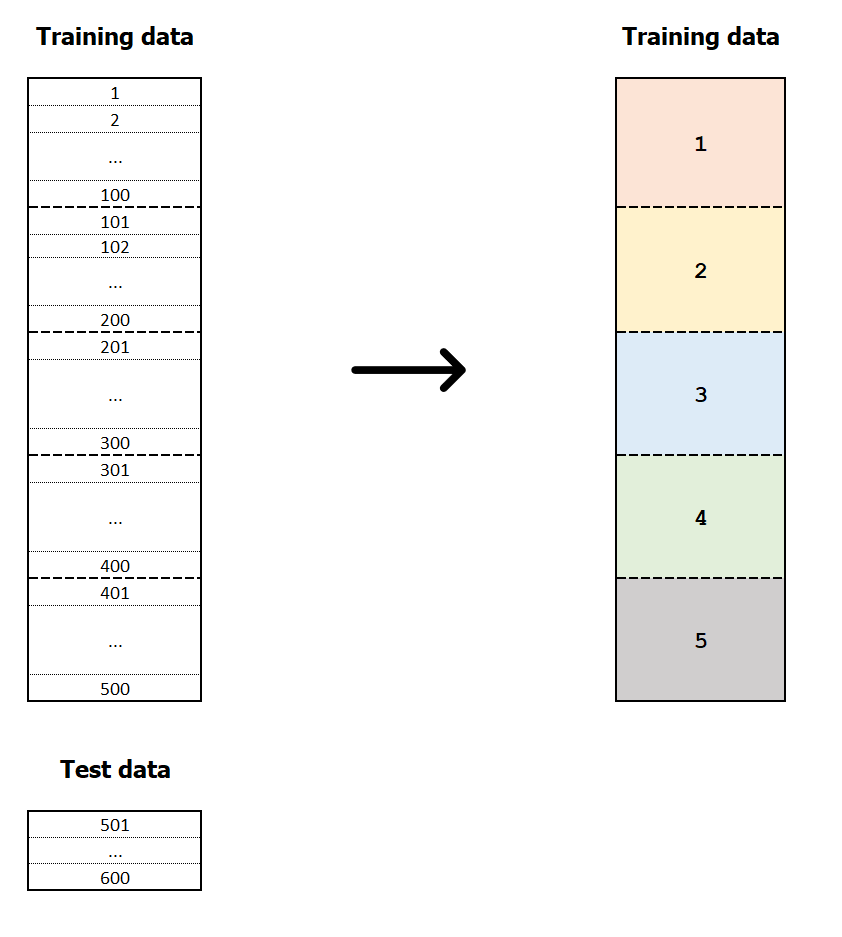
k-fold cross-validation

k-fold cross-validation
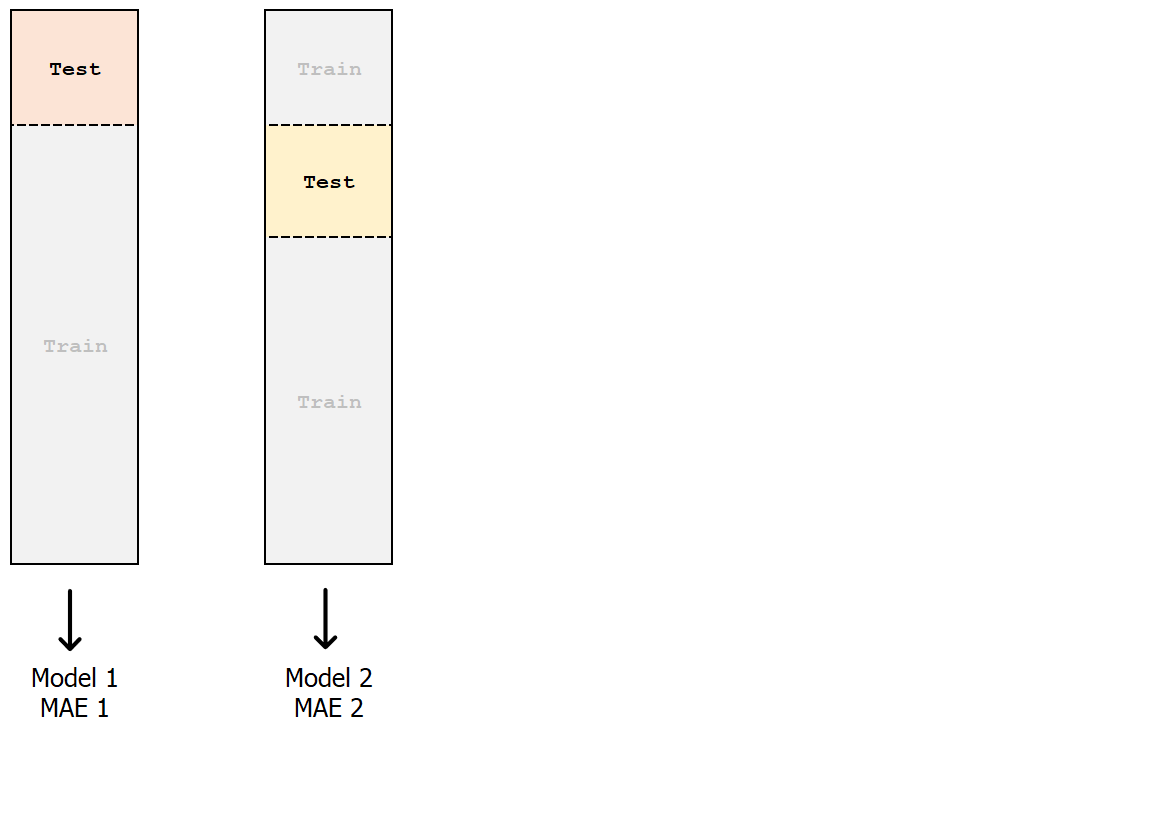
k-fold cross-validation
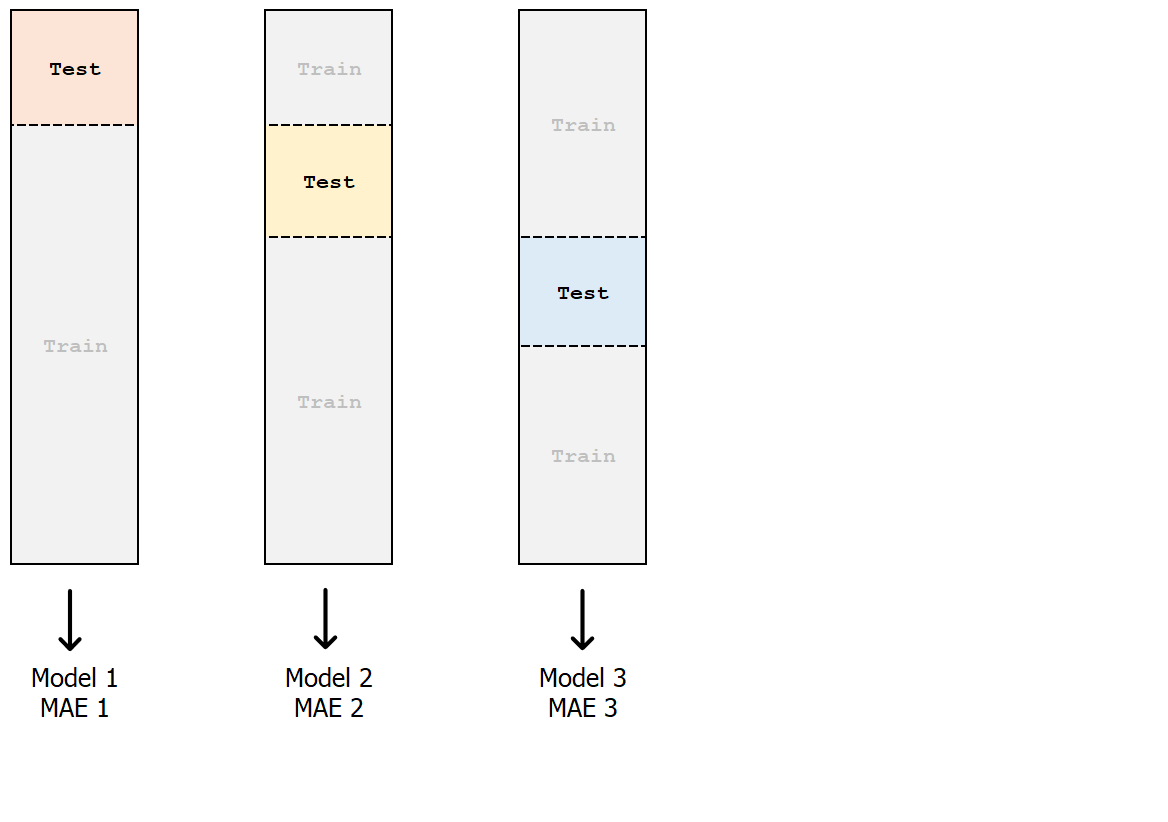
k-fold cross-validation
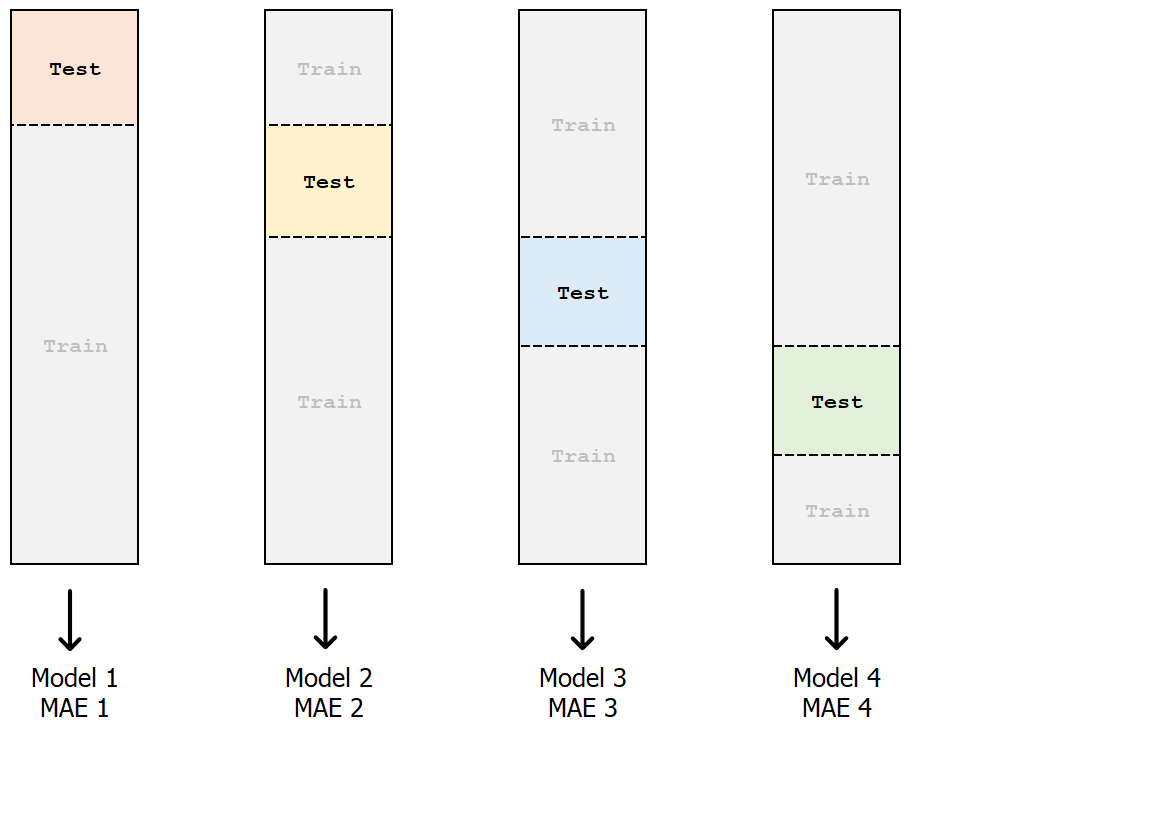
k-fold cross-validation
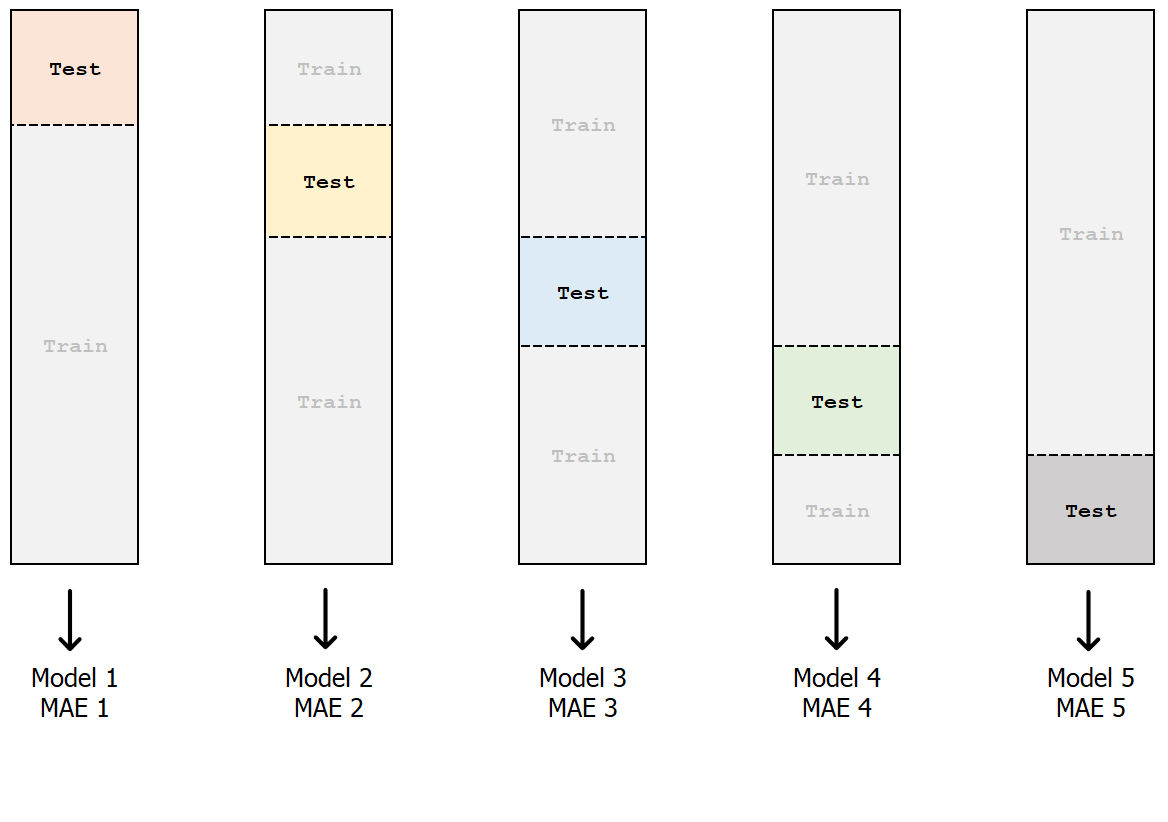
k-fold cross-validation
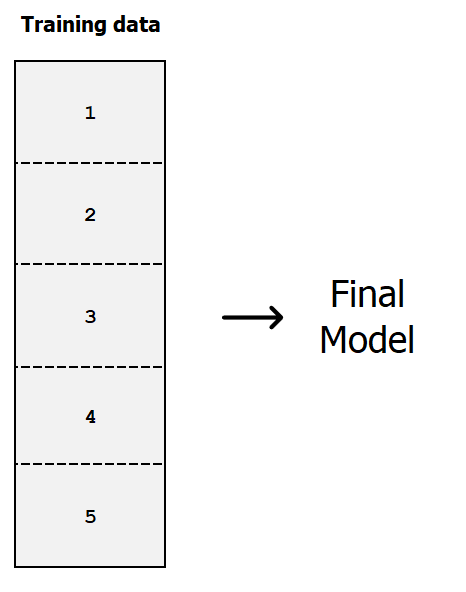
Fit final model on the full dataset
Coding - Split the data 10 times
# Random seed for reproducibility
set.seed(100)
# Create 10 folds of the dataset
chocolate_folds <- vfold_cv(chocolate_train, v = 10)
# 10-fold cross-validation
# A tibble: 10 x 2
splits id
1 <split [1293/144]> Fold1
2 <split [1293/144]> Fold2
3 <split [1293/144]> Fold3
4 ...
Coding - Fit the folds
# Fit a model for every fold and calculate MAE and RMSE fits_cv <- fit_resamples(tree_spec,final_grade ~ .,resamples = chocolate_folds,metrics = metric_set(mae, rmse))
# Resampling results
# 10-fold cross-validation
# A tibble: 10 x 4
splits id .metrics
<list> <chr> <list>
1 <split [1293/144]> Fold1 <tibble [2 x 4]>
2 <split [1293/144]> Fold2 <tibble [2 x 4]>
3 <split [1293/144]> Fold3 <tibble [2 x 4]>
4 ...
Coding - Collect all errors
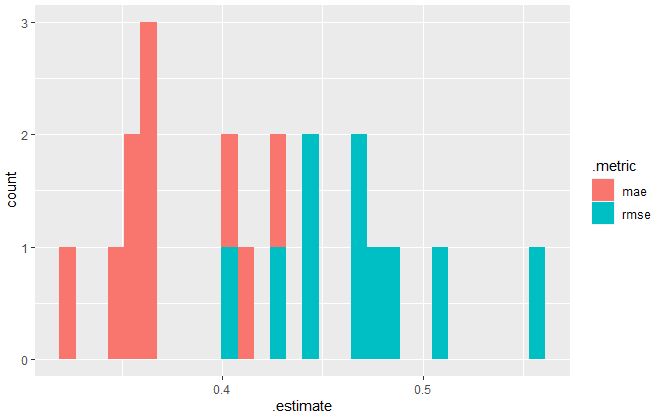
# Collect raw errors of all model runs all_errors <- collect_metrics(fits_cv, summarize = FALSE)print(all_errors)
# A tibble: 20 x 3
id .metric .estimate
<chr> <chr> <dbl>
1 Fold01 mae 0.362
2 Fold01 rmse 0.442
3 Fold02 mae 0.385
4 Fold02 rmse 0.504
5 ...
library(ggplot2)
ggplot(all_errors, aes(x = .estimate,
fill = .metric)) +
geom_histogram()
Coding - Summarize training sessions
# Collect and summarize errors of all model runs
collect_metrics(fits_cv)
# A tibble: 2 x 3
.metric mean n
<chr> <dbl> <int>
1 mae 0.383 10
2 rmse 0.477 10
Let's cross-validate!
Machine Learning with Tree-Based Models in R

