Sensitivity analysis
Monte Carlo Simulations in Python

Izzy Weber
Curriculum Manager, DataCamp
Sensitivity analysis
Helps us understand the impact of the range of inputs
Illustrates the patterns or trends when summarized in tables or plots
If we increase or decrease the values for bmi and hdl using a Monte Carlo simulation, how will the predicted y values (disease progression) change?
Defining the parameters
cov_dia = dia[["age", "bmi", "bp", "tc", "ldl", "hdl", "tch", "ltg", "glu"]].cov()
mean_dia = dia[["age", "bmi", "bp", "tc", "ldl", "hdl", "tch", "ltg", "glu"]].mean()
Defining the simulation function
def simulate_bmi_hdl(cov_dia, mean_list):list_ys = [] for i in range(50): simulation_results = st.multivariate_normal.rvs(mean=mean_list, size=500, cov=cov_dia) df_results = pd.DataFrame(simulation_results, columns=["age","bmi","bp","tc","ldl","hdl","tch","ltg","glu"]) predicted_y = regr_model.predict(df_results) df_y = pd.DataFrame(predicted_y, columns=["predicted_y"]) df_summary = pd.concat([df_results, df_y], axis=1) y = np.mean(df_summary["predicted_y"]) list_ys.append(y)return(np.mean(list_ys))
Perform simulations with a range of input parameters
hdl = [] bmi = [] simu_y = [] for mean_hdl_inc in np.arange(-20, 50, 30): for mean_bmi_inc in np.arange(-7, 11, 3):mean_list = mean_dia + np.array([0, mean_bmi_inc, 0, 0, 0, mean_hdl_inc, 0, 0, 0]) hdl.append(mean_hdl_inc) bmi.append(mean_bmi_inc)mean_y = simulate_bmi_hdl(cov_dia, mean_list)simu_y.append(mean_y)df_sa = pd.concat([pd.Series(hdl), pd.Series(bmi), pd.Series(simu_y)], axis=1)df_sa.columns = ["hdl_inc", "bmi_inc", "y"]
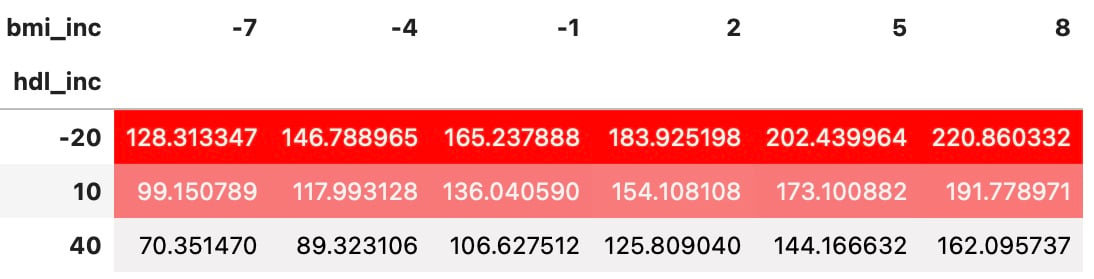
Styled DataFrames of sensitivity analysis results
df_sa.sort_values(by=['hdl_inc', 'bmi_inc']).pivot(index='hdl_inc',
columns='bmi_inc',
values='y').style.background_gradient(
cmap=sns.light_palette("red", as_cmap=True))
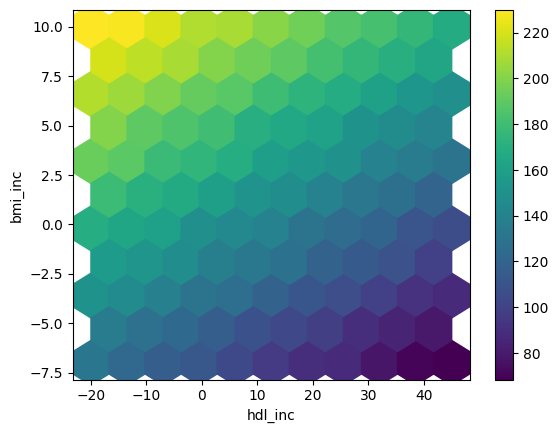
Hexbin plot for sensitivity analysis results
df_sa.plot.hexbin(x='hdl_inc',y='bmi_inc', C='y',
reduce_C_function=np.mean,
gridsize=10, cmap="viridis",
sharex=False)

Hexbin plot for dense parameter space
Let's practice!
Monte Carlo Simulations in Python

