The parallel package - parSapply
Writing Efficient R Code

Colin Gillespie
Jumping Rivers & Newcastle University
The apply family
There are parallel versions of
apply()-parApply()sapply()-parSapply()- applying a function to a vector, i.e., a for loop
lapply()-parLapply()- applying a function to a list
The sapply() function
sapply() is just another way of writing a for loop
The loop
for(i in 1:10)
x[i] <- simulate(i)
Can be written as
sapply(1:10, simulate)
We are applying a function to each value of a vector
Switching to parSapply()
It's the same recipe!
- Load the package
- Make a cluster
- Switch to
parSapply() - Stop!
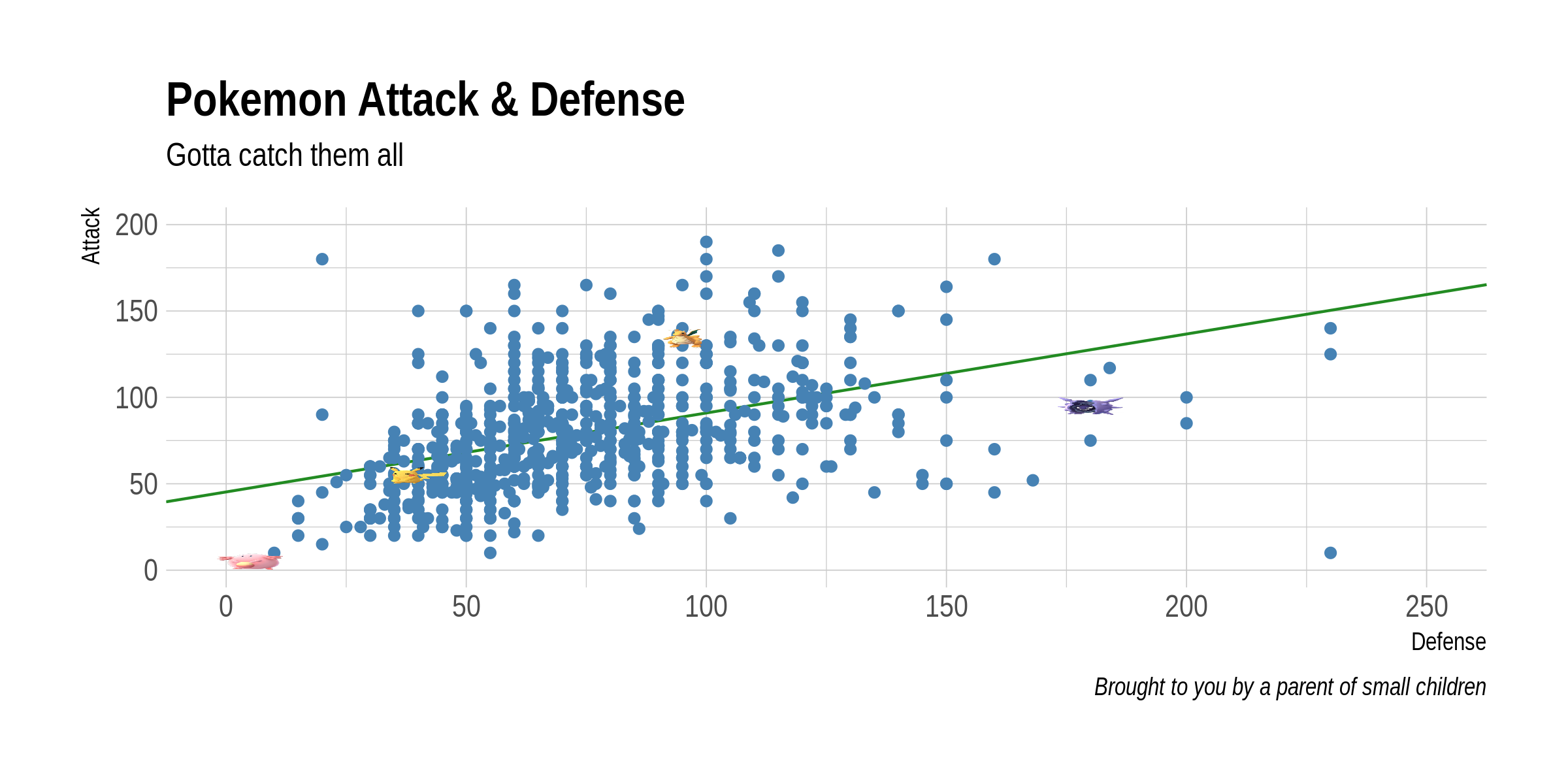
Example: Pokemon battles
plot(pokemon$Defense, pokemon$Attack)
abline(lm(pokemon$Attack ~ pokemon$Defense), col = 2)
cor(pokemon$Attack, pokemon$Defense)
0.437
Bootstrapping
In a perfect world, we would resample from the population; but we can't
Instead, we assume the original sample is representative of the population
- Sample with replacement from your data
- The same point could appear multiple times
- Calculate the correlation statistics from your new sample
- Repeat
A single bootstrap
bootstrap <- function(data_set) {
# Sample with replacement
s <- sample(1:nrow(data_set), replace = TRUE)
new_data <- data_set[s,]
# Calculate the correlation
cor(new_data$Attack, new_data$Defense)
}
# 100 independent bootstrap simulations
sapply(1:100, function(i) bootstrap(pokemon))
Converting to parallel
- Load the package
- Specify the number of cores
- Create a cluster object
- Export functions/data
- Swap to
parSapply() - Stop!
library("parallel")
no_of_cores <- 7
cl <- makeCluster(no_of_cores)
clusterExport(cl,
c("bootstrap", "pokemon"))
parSapply(cl, 1:100,
function(i) bootstrap(pokemon))
stopCluster(cl)
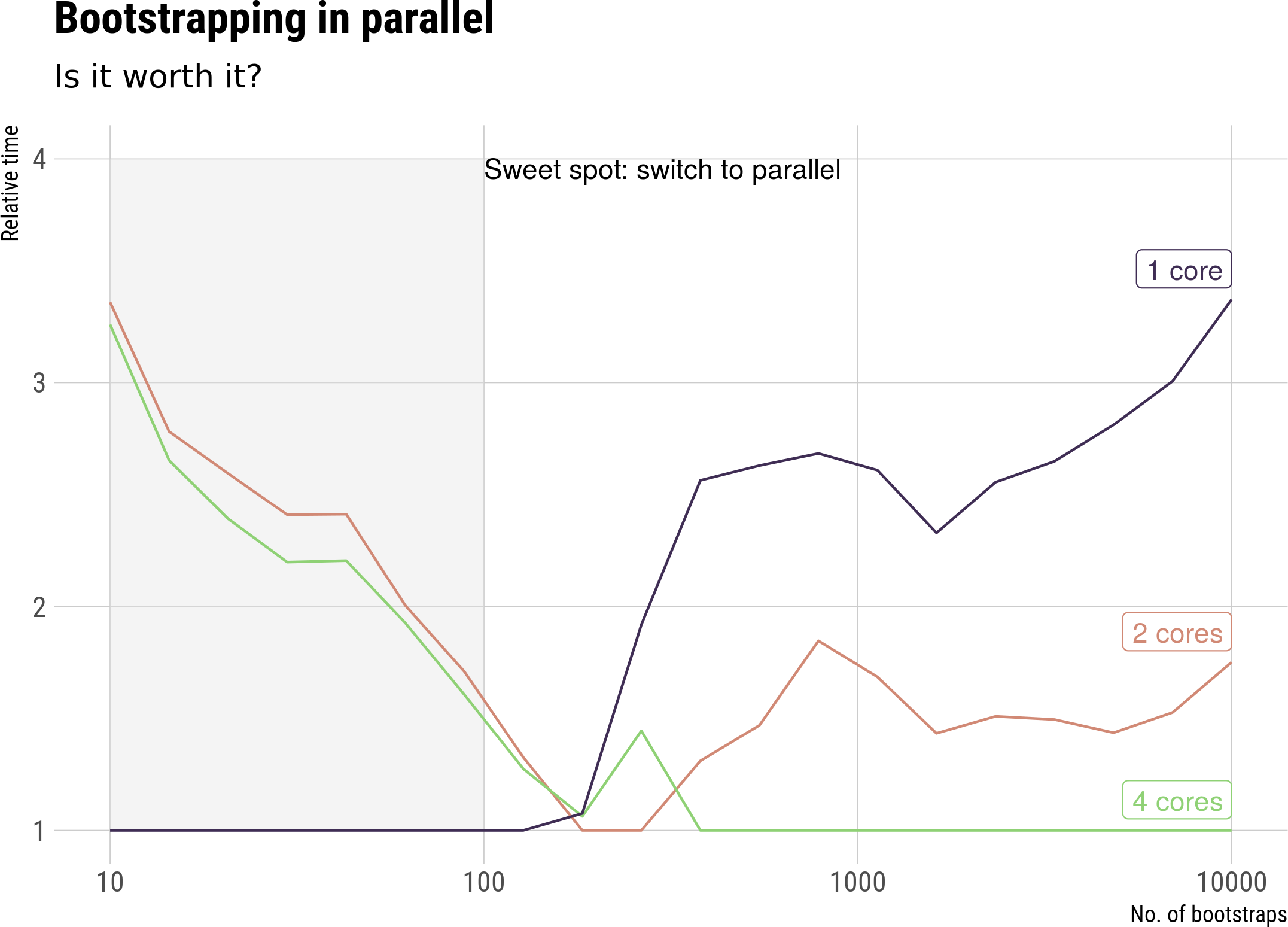
Timings
Let's practice!
Writing Efficient R Code

