Linear regression
A/B Testing in R

Lauryn Burleigh
Data Scientist
Least squares
- Residual sum of squares
- Each point residual - squared, then summed
- Line of best fit - smallest sum of squares
- Error - mean square error
- Sum of squares / N
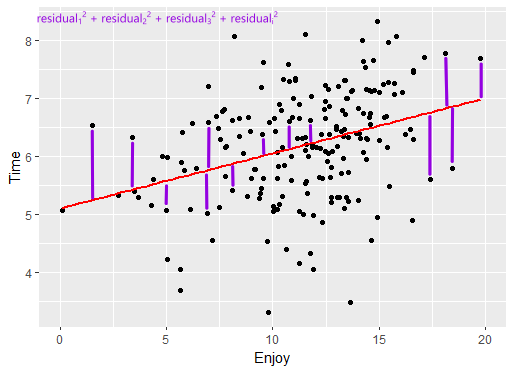
Linear regression model
linear <- lm(Time ~ Enjoy,
data = Pizza)
summary(linear)
Call:
lm(formula = Time ~ Enjoy, data = pizza)
Residuals:
Min 1Q Median 3Q Max
-2.89270 -0.59857 0.04758 0.67764 2.12600
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 5.31964 0.19886 26.750 < 2e-16 ***
Enjoy 0.07707 0.01672 4.608 7.26e-06 ***
Residual standard error: 0.8947 on 198 degrees of freedom
Multiple R-squared: 0.09687,
Adjusted R-squared: 0.09231
F-statistic: 21.24 on 1 and 198 DF, p-value: 7.262e-06
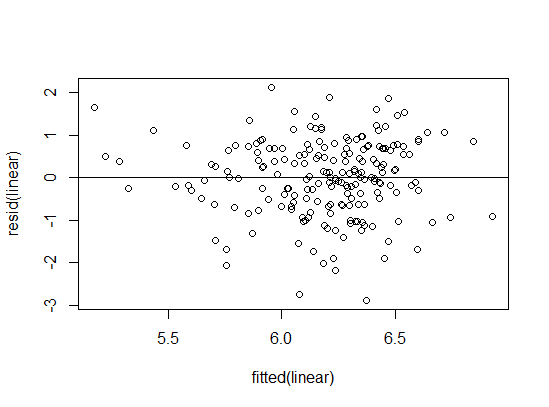
Assessing assumptions
Homoscedasticity
- Consistent variance
plot(fitted(linear), resid(linear));
abline(0,0)
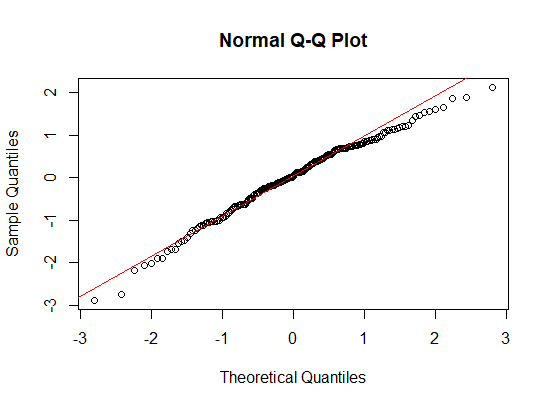
Normality
qqnorm(resid(linear));
qqline(resid(linear), col = "red")
Making predictions
Enjoy <- 12
topredict <- data.frame(Enjoy)
predict(linear, newdata = topredict)
1
6.244452
Enjoy <- c(12, 14)
topredict <- data.frame(Enjoy)
predict(linear, newdata = topredict)
1 2
6.244452 6.398587
Including groups
grplinear <- lm(Time ~ Enjoy + Topping,
data = Pizza)
summary(grplinear)
Enjoy <- c(12, 14)
Topping <- "Cheese"
topredict <- data.frame(Enjoy, Topping)
predict(grplinear, newdata = topredict)
1 2
6.136022 6.269139
Call:
lm(formula = Time ~ Enjoy + Topping,
data = pizza)
Residuals:
Min 1Q Median 3Q Max
-2.87771 -0.51529 0.03993 0.68685 2.19460
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 5.53891 0.24823 22.314 < 2e-16 ***
Enjoy 0.06656 0.01815 3.668 0.000315 ***
ToppingCheese -0.20159 0.13729 -1.468 0.143606
Residual standard error: 0.8921 on 197 degrees of freedom
Multiple R-squared: 0.1066,
Adjusted R-squared: 0.09758
F-statistic: 11.76 on 2 and 197 DF, p-value: 1.499e-05
Let's practice!
A/B Testing in R

