Measuring the benefits
Parallel Programming in R

Nabeel Imam
Data Scientist
Toy example
numbers <- 1:1000000# Sequential sqroots <- lapply(numbers, sqrt)# Parallel cl <- makeCluster(4) sqroots <- parLapply(cl, numbers, sqrt) stopCluster(my_cluster)
Which will perform better?
Benchmarking performance
Run code several times to estimate average execution time
library(microbenchmark)microbenchmark( "Sequential" = lapply(numbers, sqrt),"Parallel" = { cl <- makeCluster(4) parLapply(cl, numbers, sqrt) stopCluster(my_cluster) },times = 10 )
Unit: milliseconds
expr min mean max neval
Sequential 633.96 838.09 993.59 10
Parallel 1136.95 1247.29 1557.58 10
- Simple numerical operations rarely benefit from parallelization
- Profiling gives line-by-line report, benchmarking gives overall execution times
The elephant in the room
sqroots <- sqrt(numbers)

Vectorization
sqroots <- sqrt(numbers)
- Base R functions, like
sqrt(), are vectorized. - Map a single function to many inputs
- Very fast but only applicable to simple operations
microbenchmark(
"Vectorized" = sqrt(numbers),
"Sequential" = lapply(numbers, sqrt),
"Parallel" = {
cl <- makeCluster(4)
parLapply(cl, numbers, sqrt)
stopCluster(my_cluster)
},
times = 10)
Unit: milliseconds
expr min mean max neval
Vectorized 2.3904 9.2071 66.303 10
Sequential 352.1166 771.7491 1004.753 10
Parallel 1191.3176 1377.6926 1700.316 10
The bootstrap
Sampling from the current data with replacement
print(ls_df)
$`2001`
Country Life_expectancy Year
1 Afghanistan 56.3 2001
2 Albania 74.3 2001
3 Algeria 71.1 2001
...
$`2002`
Country Life_expectancy Year
1 Afghanistan 56.8 2002
2 Albania 74.6 2002
3 Algeria 71.6 2002
...
Classic version
df <- ls_df$`2001`estimates <- rep(0, 10000)for (i in 1:10000) { b <- sample(df$Life_expectancy, replace = T)estimates[i] <- mean(b) }
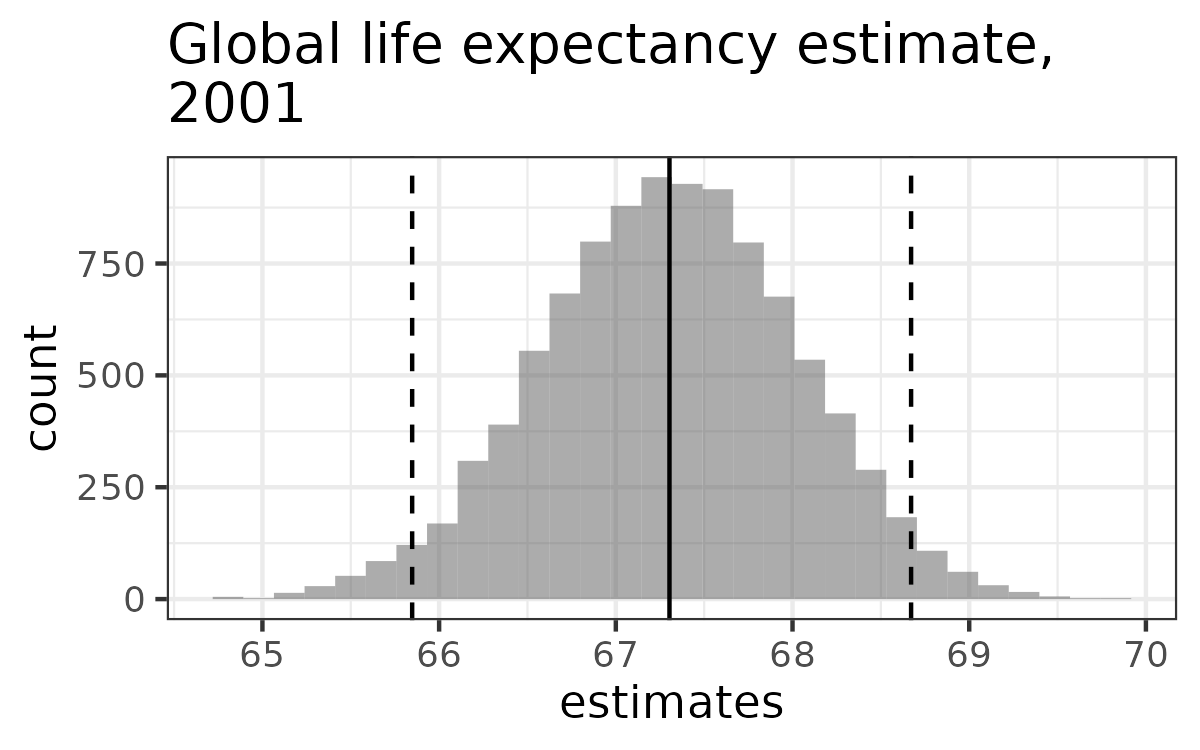
- Confidence interval using quantiles:
quantile(estimates, c(0.025, 0.975))
The good news
Bootstraps can be parallelized
estimates <- rep(0, 10000)
for (i in 1:10000) {
b <- sample(df$Life_expectancy,
replace = T)
estimates[i] <- mean(b)
}
boot_dist <- function (df) { estimates <- rep(0, 10000) for (i in 1:10000) { b <- sample(df$Life_expectancy, replace = T) estimates[i] <- mean(b) } return(estimates) }cl <- makeCluster(4) ls_dists <- parLapply(cl, ls_df, boot_dist) stopCluster(cl)
The benefits
microbenchmark(
"lapply" = lapply(ls_df, boot_dist),
"parLapply" = {
cl <- makeCluster(4)
parLapply(cl, ls_df, boot_dist)
stopCluster(cl)
},
times = 10
)
Unit: seconds
expr min mean max neval
lapply 3.6938 4.2184 4.5267 10
parLapply 1.9006 2.5166 2.7292 10
How to get there:
- Profile existing code, identify slowest part
- Parallelize/optimize this step
- Benchmark and compare
Let's practice!
Parallel Programming in R

