Monitoring and managing memory
Parallel Programming in R

Nabeel Imam
Data Scientist
The queue and the space

The parallel flow
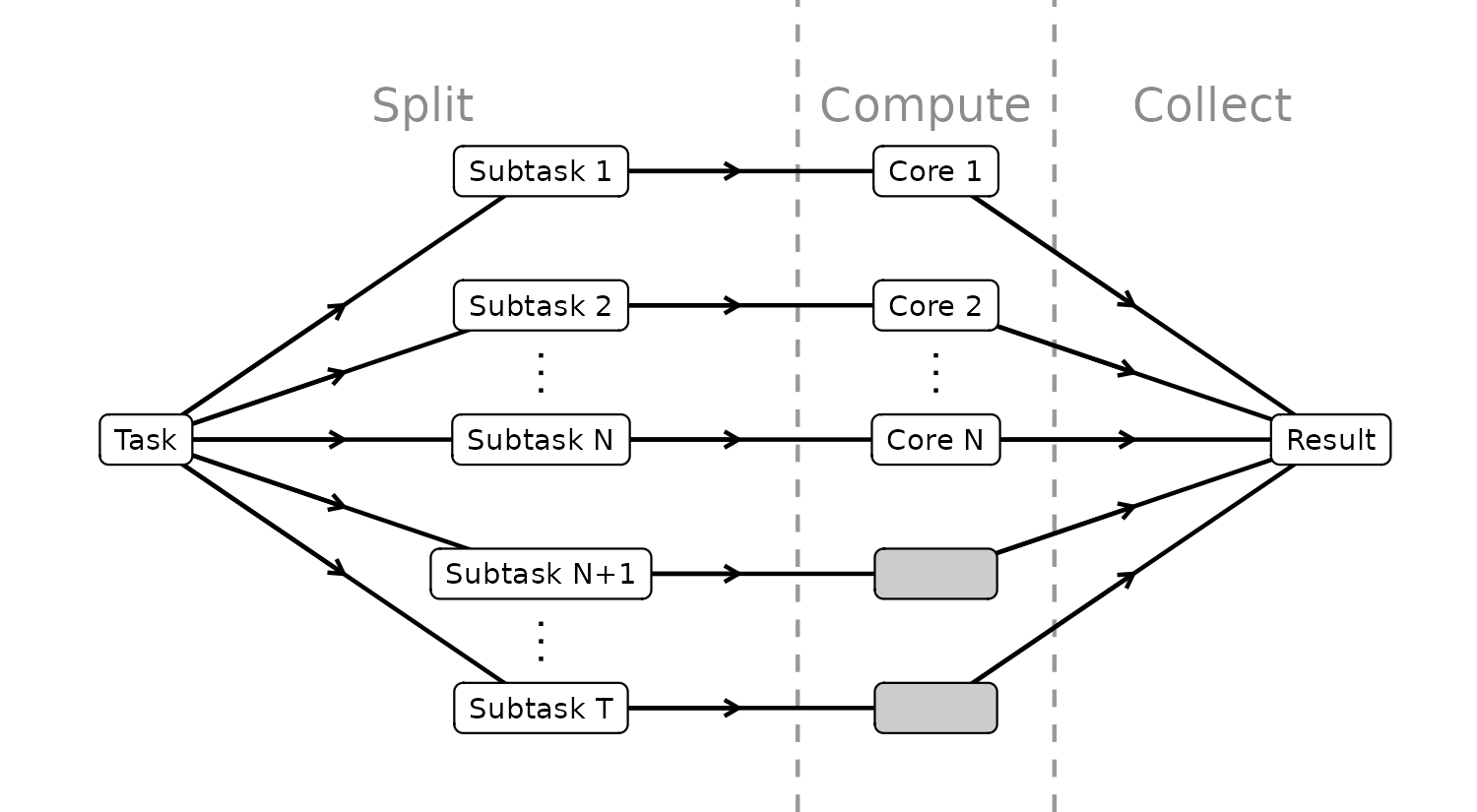
The parallel flow
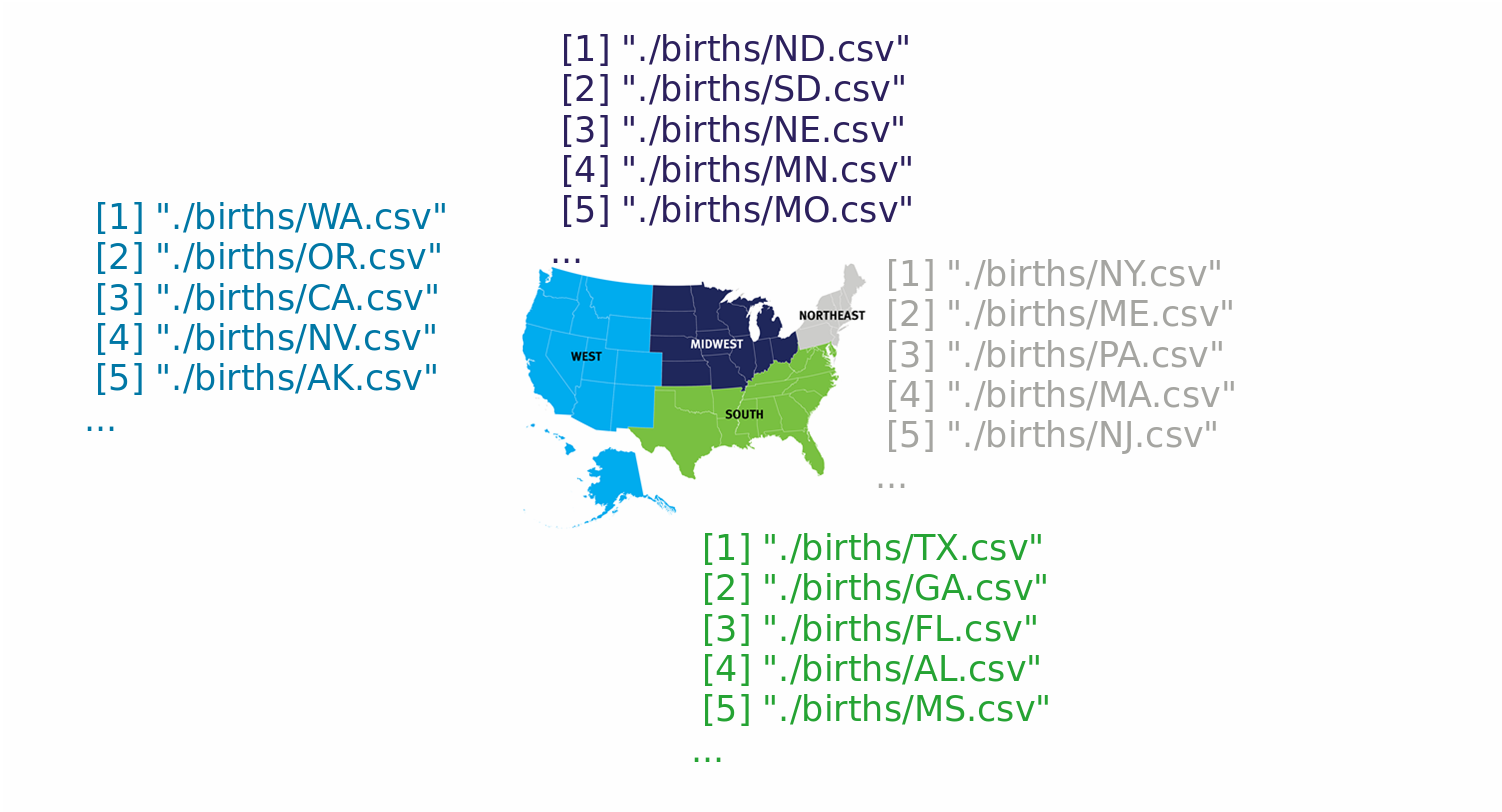
The births data
print(ls_files)
[1] "./births/AK.csv"
[2] "./births/AL.csv"
[3] "./births/AR.csv"
[4] "./births/AZ.csv"
[5] "./births/CA.csv"
[6] "./births/CO.csv"
[7] "./births/CT.csv"
[8] "./births/DC.csv"
[9] "./births/DE.csv"
[10] "./births/FL.csv"
...
Mapping with futures
plan(multisession, workers = 2)ls_df <- future_map(ls_files, read.csv)plan(sequential)print(ls_df)
[[1]]
state month plurality weight_gain_pounds mother_age
AK 1 1 30 43
...
[[2]]
state month plurality weight_gain_pounds mother_age
AL 10 1 60 33
...
...
Profiling with two workers
profvis({
plan(multisession, workers = 2)
ls_df <- future_map(ls_files, read.csv)
plan(sequential)
})

Profiling with four workers
profvis({
plan(multisession, workers = 4)
ls_df <- future_map(ls_files, read.csv)
plan(sequential)
})
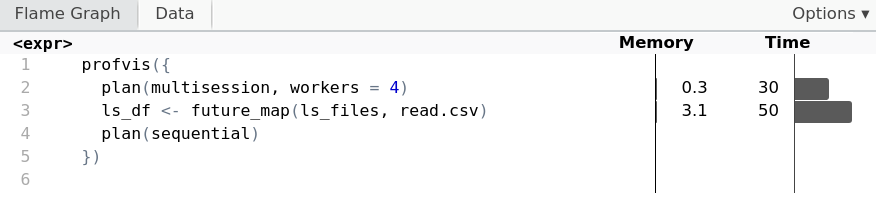
Behind the scenes
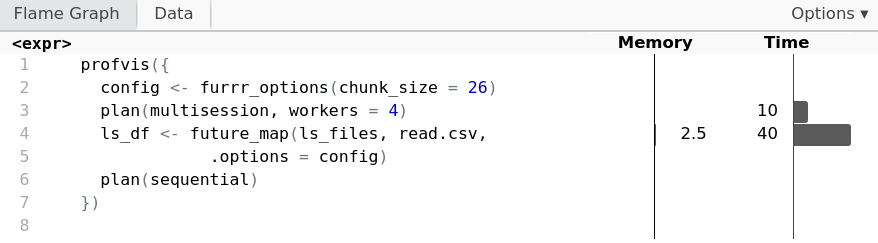
Managing memory by chunking
config <- furrr_options(chunk_size = 26)plan(multisession, workers = 4) ls_df <- future_map(ls_files, read.csv,.options = config) plan(sequential)
Managing memory by chunking
profvis({
config <- furrr_options(chunk_size = 26)
plan(multisession, workers = 4)
ls_df <- future_map(ls_files, read.csv,
.options = config)
plan(sequential)
})
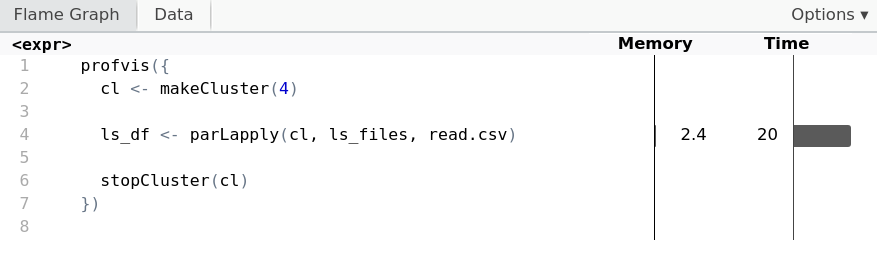
Chunking with parallel
cl <- makeCluster(4)ls_df <- parLapply(cl, ls_files, read.csv)stopCluster(cl)
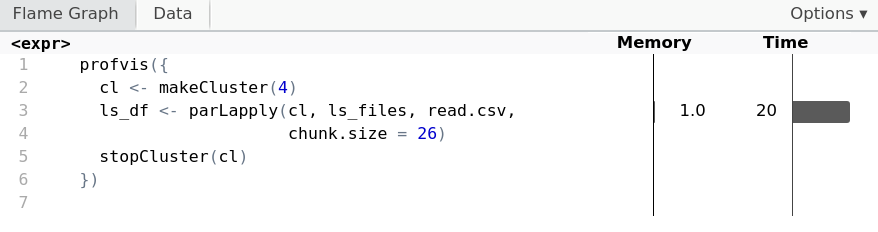
Chunking with parallel
cl <- makeCluster(4) ls_df <- parLapply(cl, ls_files, read.csv,chunk.size = 26)stopCluster(cl)
When to chunk?
- Chunking is performed optimally by default
- With large data objects and running low on memory
- Try using fewer cores if feasible
- Experiment with a few chunk sizes to get to optimum
Let's practice!
Parallel Programming in R

