Debugging in parallel
Parallel Programming in R

Nabeel Imam
Data Scientist
What is debugging?

Reading files in parallel
print(file_list)
[1] "./stocks/2011.csv"
[2] "./stocks/2012.csv"
[3] "./stocks/2013.csv"
[4] "./stocks/2014.csv"
[5] "./stocks/2015.csv"
...
The filtering function
filterCSV <- function (filepath) {
# Read CSV
df <- read.csv(filepath)
# Filter data
df <- df %>%
dplyr::filter(Company == "Tesla")
# Write to back to same path
write.csv(df, filepath)
}
The parallel apply
cl <- makeCluster(4)clusterEvalQ(cl, library(dplyr))dummy <- parLapply(cl, file_list, filterCSV)stopCluster(cl)
Error in checkForRemoteErrors(val) :
one node produced an error: ? In argument: `Company == "Tesla"`.
Caused by error:
! object 'Company' not found
The sequential run
short_list <- file_list[1:5]dummy <- lapply(short_list, filterCSV)read.csv(short_list[1])
Date Open High Low Close Adj.Close Volume Company Year
1 2011-01-03 5.368 5.400 5.180 5.324 5.324 6415000 Tesla 2011
2 2011-01-04 5.332 5.390 5.204 5.334 5.334 5937000 Tesla 2011
3 2011-01-05 5.296 5.380 5.238 5.366 5.366 7233500 Tesla 2011
...
Locate the error
Error message
Error in checkForRemoteErrors(val) :
one node produced an error:
In argument: `Company == "Tesla"`.
Caused by error:
! object 'Company' not found

Locate the error
filterCSV <- function (filepath) {
# Read CSV
df <- read.csv(filepath)
# Filter data
df <- df %>%
dplyr::filter(Company == "Tesla")
# Write to back to same path
write.csv(df, filepath)
}
filterCSV_debug <- function (filepath) { df <- read.csv(filepath)print(# Paste file path and column names paste(filepath, ":",# Collapse column names into one string paste0(colnames(df), collapse = ",")))df <- df %>% dplyr::filter(Company == "Tesla") write.csv(df, filepath) }
Locate the error
cl <- makeCluster(4)
clusterEvalQ(cl, library(dplyr))
dummy <- parLapply(cl, file_list, filterCSV_debug)
stopCluster(cl)
Error in checkForRemoteErrors(val) :
one node produced an error: ? In argument: `Company == "Microsoft"`.
Caused by error:
! object 'Company' not found
Locate the error
cl <- makeCluster(4, outfile = "log.txt") # Log print messages into "log.txt"clusterEvalQ(cl, library(dplyr)) parLapply(cl, file_list, filterCSV_debug) stopCluster(cl)
Error in checkForRemoteErrors(val) :
one node produced an error: ? In argument: `Company == "Tesla"`.
Caused by error:
! object 'Company' not found
Examining logs
Debugging with foreach
cl <- makeCluster(4,
# Supply a text file name to log print messages
outfile = "log.txt")
registerDoParallel(cl)
foreach(f = file_list,
.packages = "dplyr") %dopar% {
filterCSV_debug(f)
}
stopCluster(cl)
The good thing about furrr
plan(multisession, workers = 4)
future_map(file_list, filterCSV_debug)
plan(sequential)
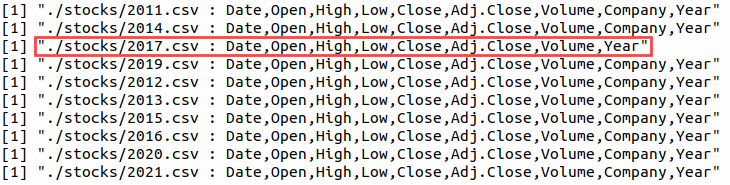
The good thing about furrr
[1] "./stocks/2011.csv : Date,Open,High,Low,Close,Adj.Close,Volume,Company,Year"
[1] "./stocks/2012.csv : Date,Open,High,Low,Close,Adj.Close,Volume,Company,Year"
[1] "./stocks/2013.csv : Date,Open,High,Low,Close,Adj.Close,Volume,Company,Year"
[1] "./stocks/2014.csv : Date,Open,High,Low,Close,Adj.Close,Volume,Company,Year"
[1] "./stocks/2015.csv : Date,Open,High,Low,Close,Adj.Close,Volume,Company,Year"
[1] "./stocks/2016.csv : Date,Open,High,Low,Close,Adj.Close,Volume,Company,Year"
[1] "./stocks/2017.csv : Date,Open,High,Low,Close,Adj.Close,Volume,Year"
Error in (function (.x, .f, ..., .progress = FALSE) :
? In index: 1.
Caused by error in `dplyr::filter()`:
? In argument: `Company == "Tesla"`.
Caused by error:
! object 'Company' not found
The steps
For errors in parallel
- Do a sequential run on a subset of the input
- Examine the error message and print appropriate messages
- Locate the error by printing or logging messages
- Fix the error
Let's practice!
Parallel Programming in R

