Parallelization in R
Parallel Programming in R

Nabeel Imam
Data Scientist
A practical example
The data
print(file_list)
[1] "./uni_data_country/Argentina.csv"
[2] "./uni_data_country/Armenia.csv"
[3] "./uni_data_country/Australia.csv"
[4] "./uni_data_country/Austria.csv"
[5] "./uni_data_country/Azerbaijan.csv"
[6] "./uni_data_country/Bahrain.csv"
[7] "./uni_data_country/Bangladesh.csv"
[8] "./uni_data_country/Belarus.csv"
[9] "./uni_data_country/Belgium.csv"
[10] "./uni_data_country/Bolivia.csv"
...

Add a column
for (file in file_list) { df <- read.csv(file)df$top100 <- NA for (r in 1:nrow(df)) { df$top100[r] <- df$world_rank[r] <= 100 }write.csv(df, file) }
Profiling
Code
library(profvis)profvis({for (file in file_list) { df <- read.csv(file) df$top100 <- NA for (r in 1:nrow(df)) { df$top100[r] <- df$Rank[r] <= 100 } write.csv(df, file) }})
Output
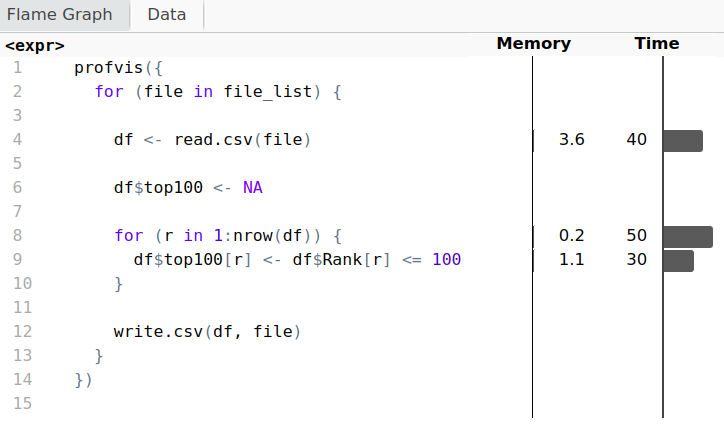
Let's parallelize
The loop
for (file in file_list) {
df <- read.csv(file)
df$top100 <- NA
for (r in 1:nrow(df)) {
df$top100[r] <- df$Rank[r] <= 100
}
write.csv(df, file)
}
Function
add_col <- function(file_path) { df <- read.csv(file_path) df$top100 <- NA for (r in 1:nrow(df)) { df$top100[r] <- df$Rank[r] <= 100 } write.csv(df, file_path) }cl <- makeCluster(6)dummy <- parLapply(cl, file_list, add_col) stopCluster(cl)
Practical considerations: number of cores
Detecting cores
detectCores()
[1] 8
Parallelized code
cl <- makeCluster(detectCores() - 2)dummy <- parLapply(cl, file_list, add_col) stopCluster(cl)
Practical considerations: cluster type
PSOCK cluster (default)
cl <- makeCluster(detectCores() - 2)
- Creates copies of current R session
- Cores do not share memory
- Works on any OS (Windows, Mac, Linux)
FORK cluster
cl <- makeCluster(detectCores() - 2,
type = "FORK")
- Creates subprocesses from R session
- Cores share memory (faster than PSOCK)
- Does not work on Windows
Let's exercise!
Parallel Programming in R

