Measures of spread
Introduction to Statistics in R

Maggie Matsui
Content Developer, DataCamp
What is spread?
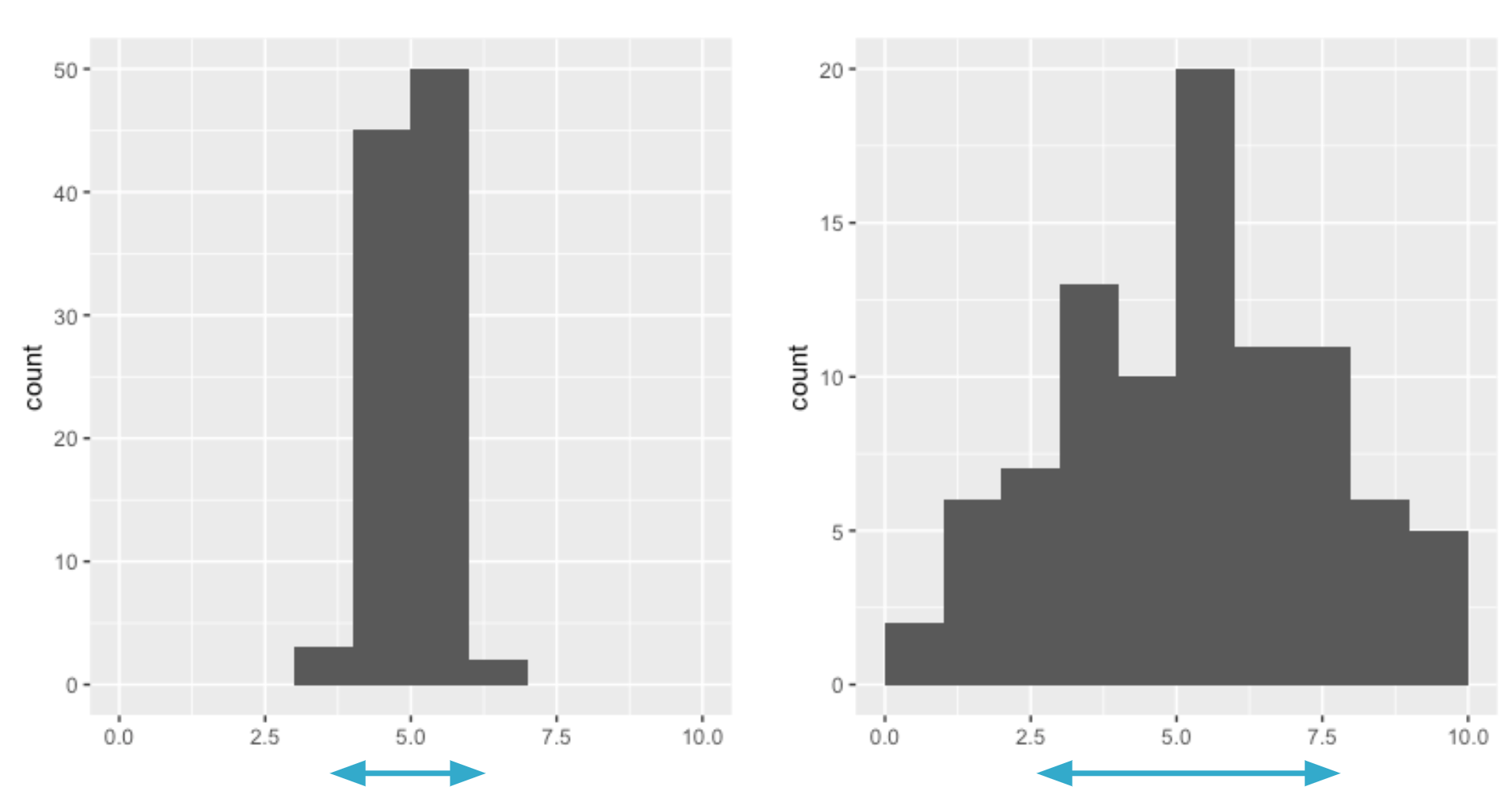
Variance
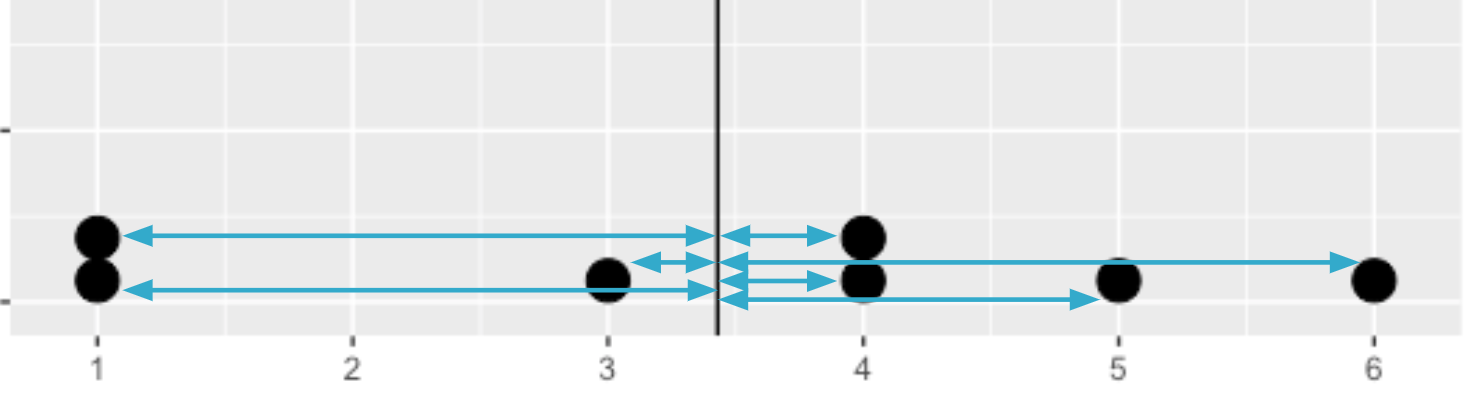
Average distance from each data point to the data's mean
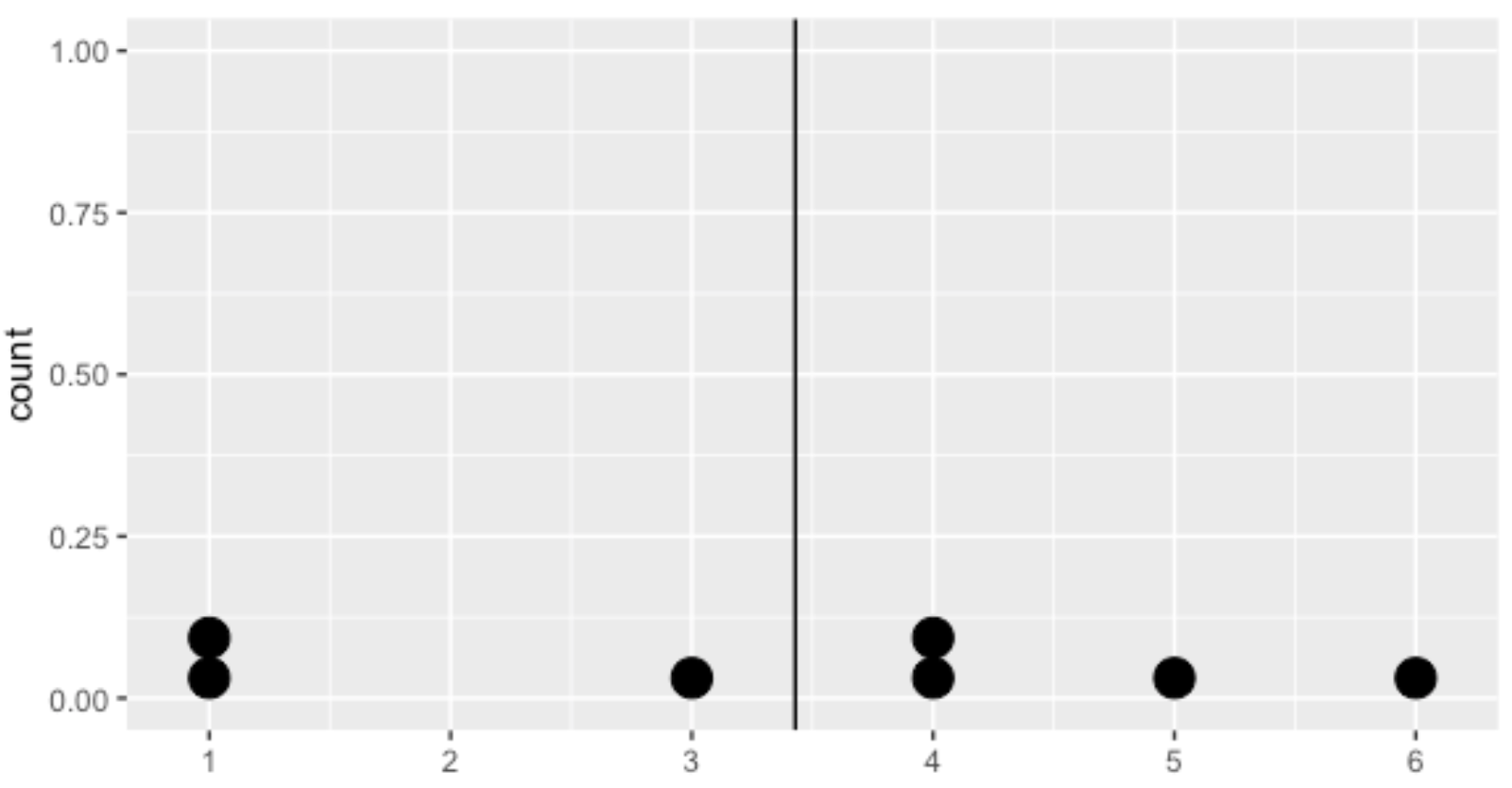
Calculating the variance
dists <- msleep$sleep_total - mean(msleep$sleep_total)
dists
1.66626506 6.56626506 ... -4.13373494 2.06626506 -0.63373494
Calculating the variance
squared_dists <- (dists)^2
2.776439251 43.115836841 ... 17.087764552 4.269451299 0.401619974
sum_sq_dists <- sum(squared_dists)
sum_sq_dists
1624.066
Calculating the variance
sum_sq_dists/82
19.80568
var(msleep$sleep_total)
19.80568
Standard deviation
sqrt(var(msleep$sleep_total))
4.450357
# Standard deviation of 'sleep_total'
sd(msleep$sleep_total)
4.450357
Mean absolute deviation
dists <- msleep$sleep_total - mean(msleep$sleep_total)
mean(abs(dists))
3.566701
Standard deviation vs. mean absolute deviation
- SD squares distances, penalizing longer distances more than shorter ones.
- MAD penalizes each distance equally.
- One isn't better than the other, but SD is more common than MAD.
Quartiles
quantile(msleep$sleep_total)
0% 25% 50% 75% 100%
1.90 7.85 10.10 13.75 19.90
Second quartile/50th percentile = median
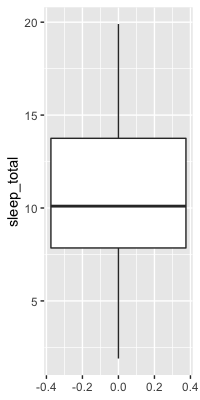
Boxplots use quartiles
ggplot(msleep, aes(y = sleep_total)) +
geom_boxplot()
Quantiles
quantile(msleep$sleep_total, probs = c(0, 0.2, 0.4, 0.6, 0.8, 1))
0% 20% 40% 60% 80% 100%
1.90 6.24 9.48 11.14 14.40 19.90
seq(from, to, by)
quantile(msleep$sleep_total, probs = seq(0, 1, 0.2))
0% 20% 40% 60% 80% 100%
1.90 6.24 9.48 11.14 14.40 19.90
Interquartile range (IQR)
Height of the box in a boxplot
iqr = quantile(msleep$sleep_total, 0.75) - quantile(msleep$sleep_total, 0.25)
iqr
75%
5.9
Outliers
Outlier: data point that is substantially different from the others
How do we know what a substantial difference is? A data point is an outlier if:
- $\text{data} < \text{Q1} - 1.5\times\text{IQR}$ or
- $\text{data} > \text{Q3} + 1.5\times\text{IQR}$
Finding outliers
iqr <- quantile(msleep$bodywt, 0.75) - quantile(msleep$bodywt, 0.25)lower_threshold <- quantile(msleep$bodywt, 0.25) - 1.5 * iqr upper_threshold<- quantile(msleep$bodywt, 0.75) + 1.5 * iqr
msleep %>% filter(bodywt < lower_threshold | bodywt > upper_threshold ) %>%
select(name, vore, sleep_total, bodywt)
# A tibble: 11 x 4
name vore sleep_total bodywt
<chr> <chr> <dbl> <dbl>
1 Cow herbi 4 600
2 Asian elephant herbi 3.9 2547
3 Horse herbi 2.9 521
...
Let's practice!
Introduction to Statistics in R

