Facet labels and order
Intermediate Data Visualization with ggplot2

Rick Scavetta
Founder, Scavetta Academy
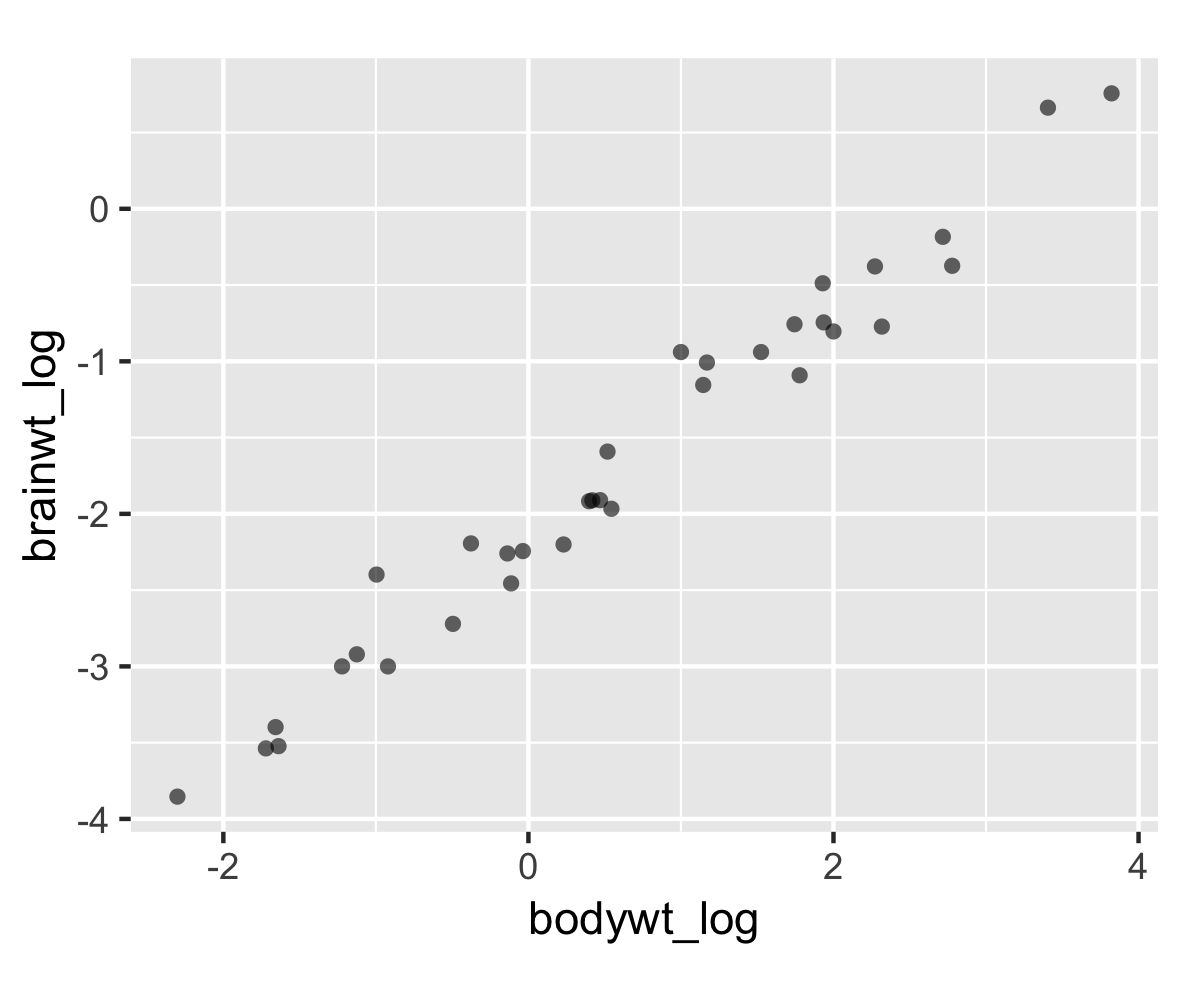
A new dataframe
# Plot
p <- ggplot(msleep2, aes(bodywt_log,
brainwt_log)) +
geom_point(alpha = 0.6, shape = 16) +
coord_fixed()
p
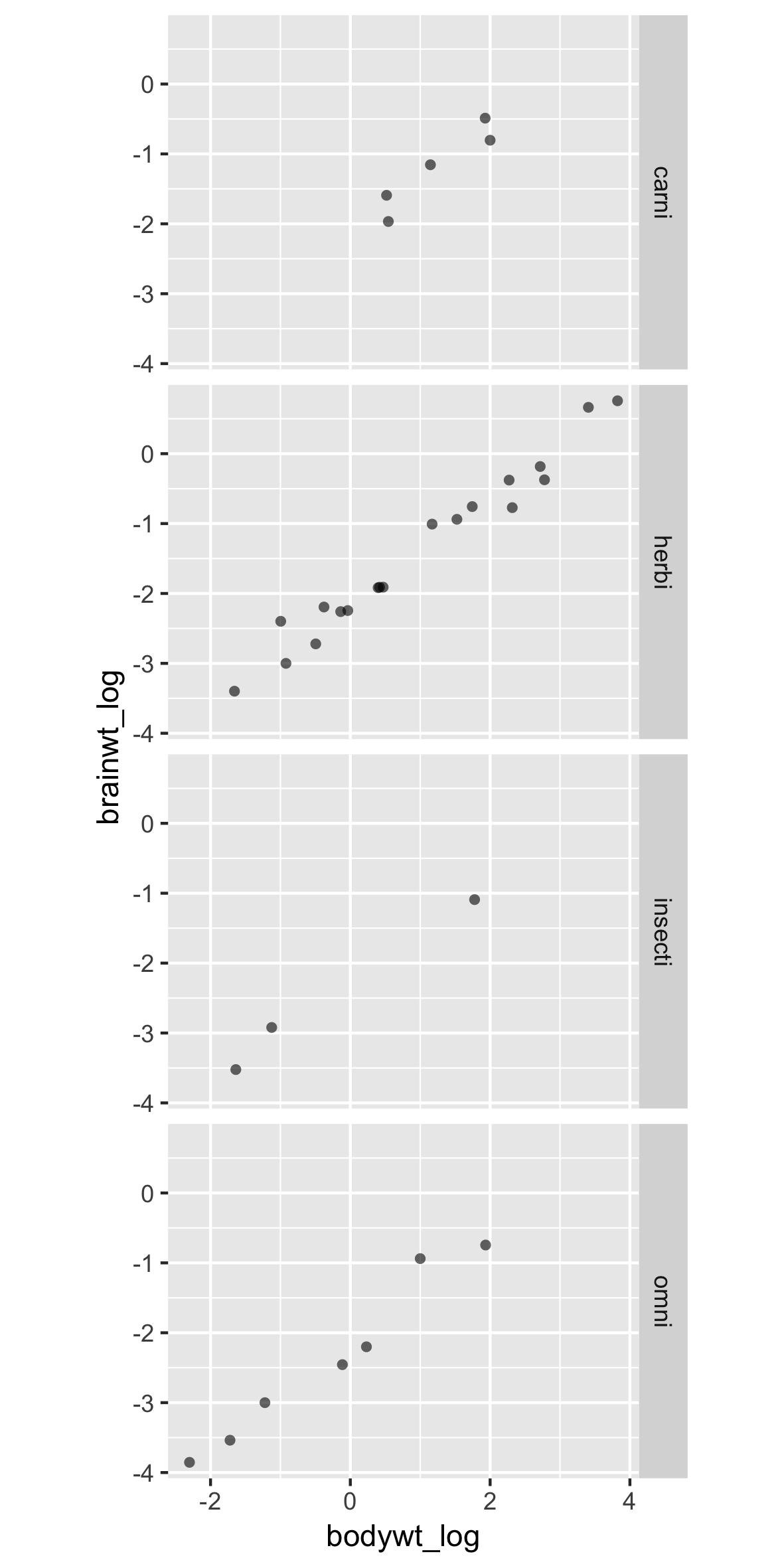
A new dataframe, with facets
p +
facet_grid(rows = vars(vore))
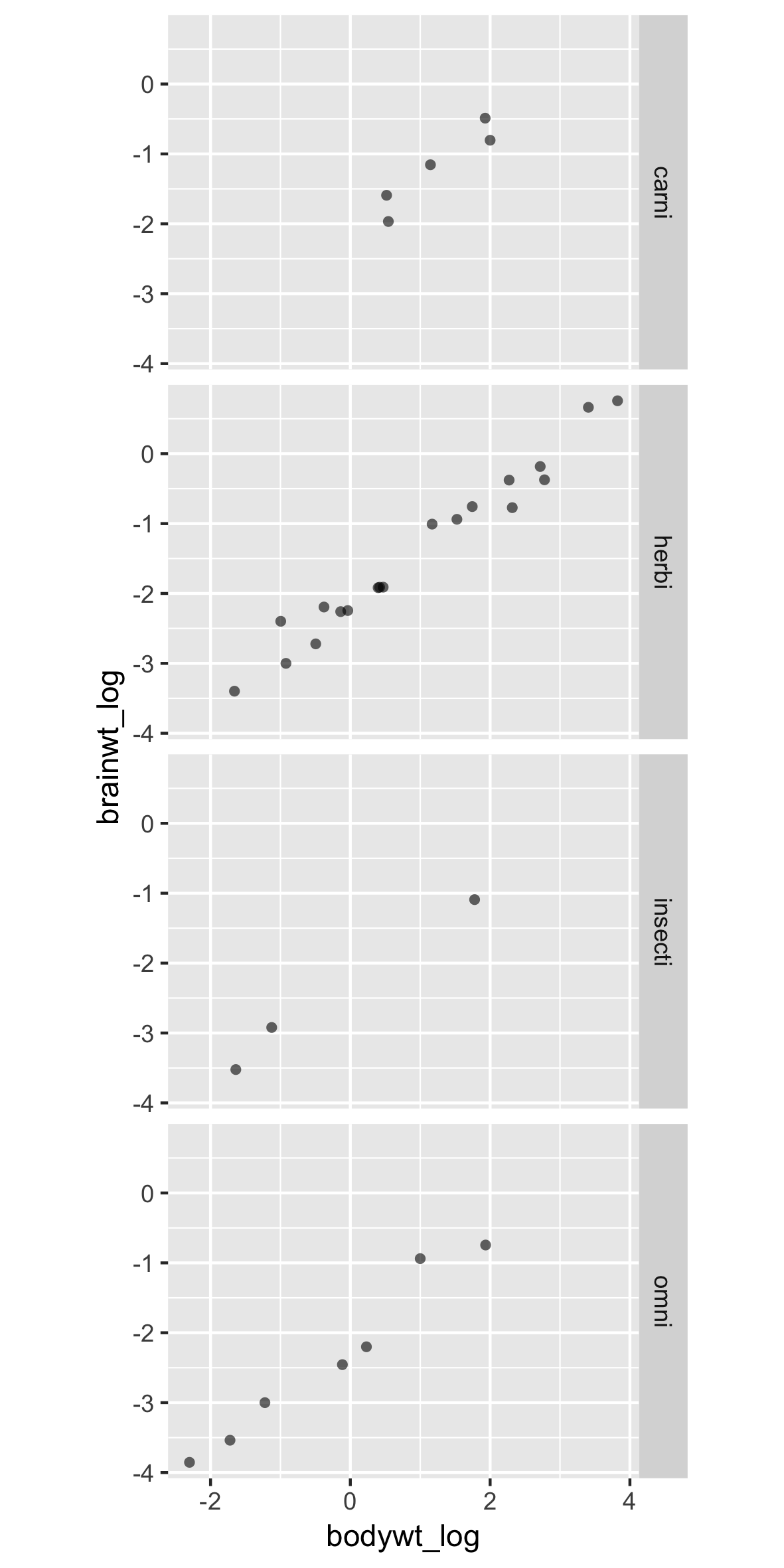
A new dataframe, with facets
p +
facet_grid(rows = vars(vore))

Poor labels and order
p +
facet_grid(rows = vars(vore))
Two typical problems with facets:
- Poorly labeled (e.g. non descriptive)
- Wrong or inappropriate order

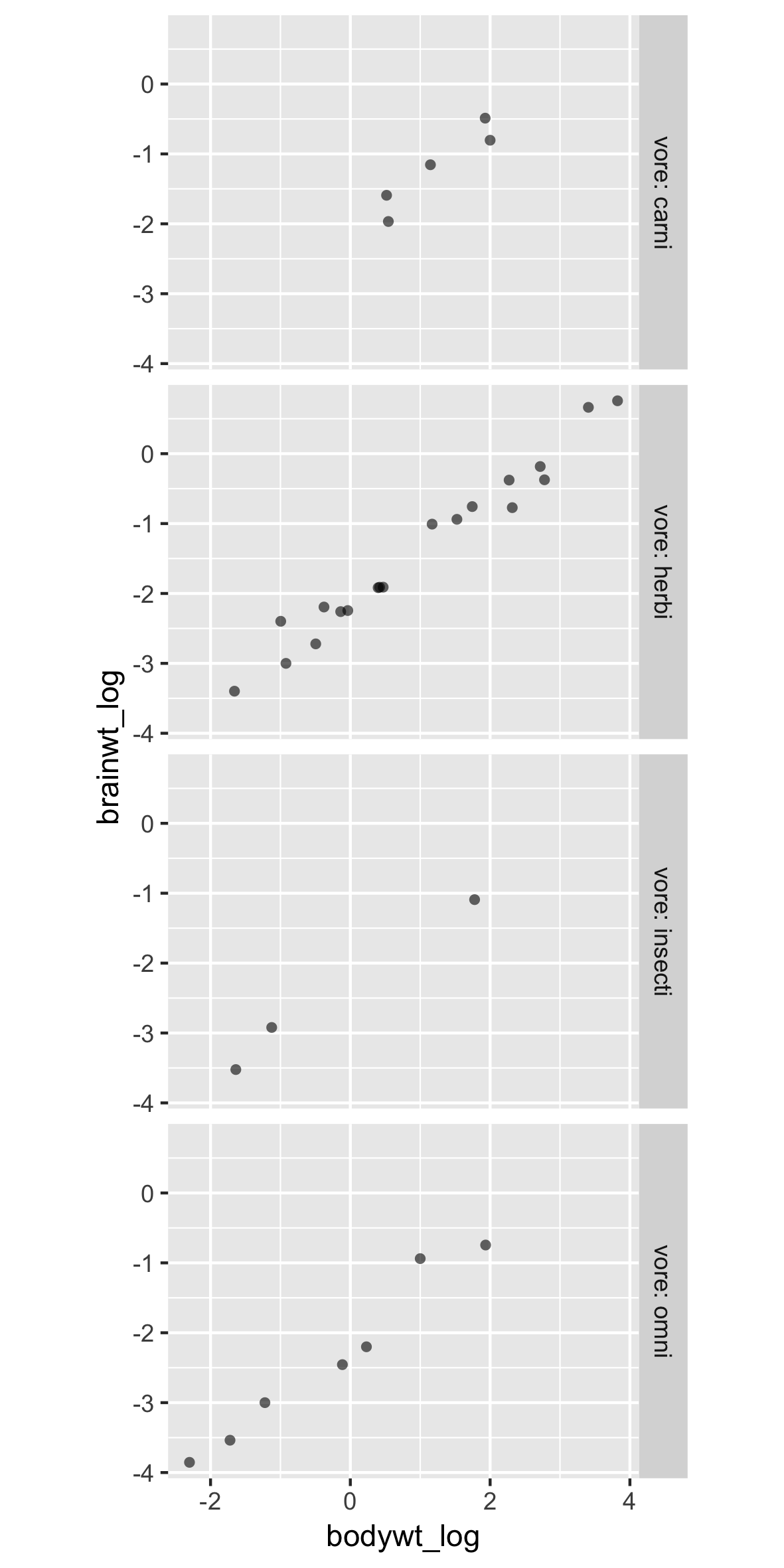
Poor labels and order
p +
facet_grid(rows = vars(vore))
Solutions:
- Easy: Add labels in ggplot
- Better: Relabel and rearrange factor variables in your dataframe

The labeller argument
# Default is to label the value
p +
facet_grid(rows = vars(vore),
labeller = label_value)
Using label_both adds the variable name
# Print variable name also
p +
facet_grid(rows = vars(vore),
labeller = label_both)
Two variables on one side
p +
facet_grid(rows = vars(vore,
conservation))

Using label_context avoids ambiguity
p +
facet_grid(rows = vars(vore,
conservation),
labeller = label_context)

Use rows and columns when appropriate
p +
facet_grid(rows = vars(vore),
cols = vars(conservation),
labeller = label_context)
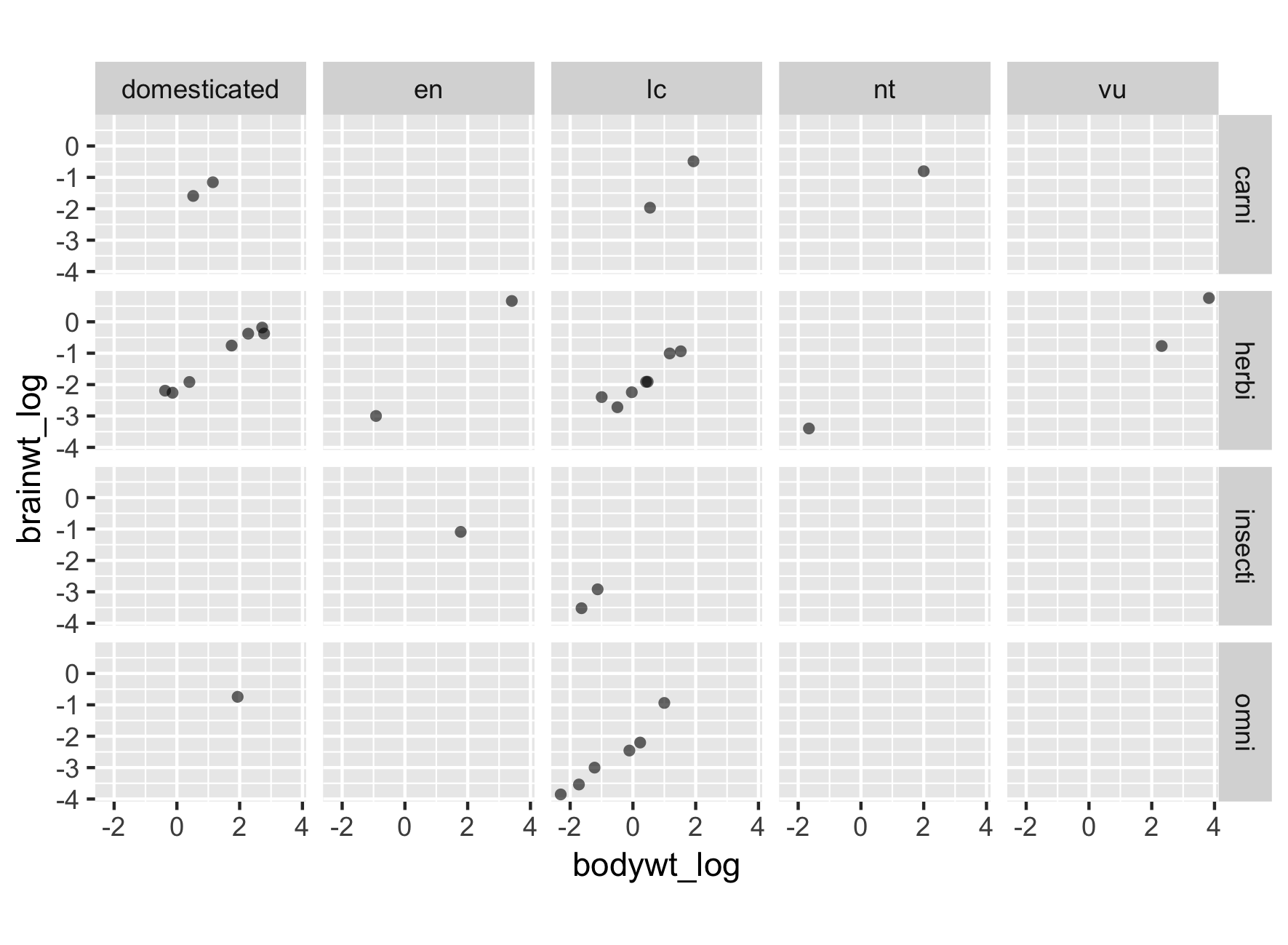
Use rows and columns when appropriate
p +
facet_grid(rows = vars(vore),
cols = vars(conservation))

Use rows and columns when appropriate

Relabeling and reordering factors
msleep2$conservation <- fct_recode(msleep2$conservation,
Domesticated = "domesticated",
`Least concern` = "lc",
`Near threatened` = "nt",
Vulnerable = "vu",
Endangered = "en")
msleep2$vore = fct_recode(msleep2$vore,
Carnivore = "carni",
Herbivore = "herbi",
Insectivore = "insecti",
Omnivore = "omni")
Reinitialize plot with new labels
# Plot
p <- ggplot(msleep2, aes(bodywt_log,
brainwt_log)) +
geom_point(alpha = 0.6, shape = 16) +
coord_fixed()
p +
facet_grid(rows = vars(vore),
cols = vars(conservation))
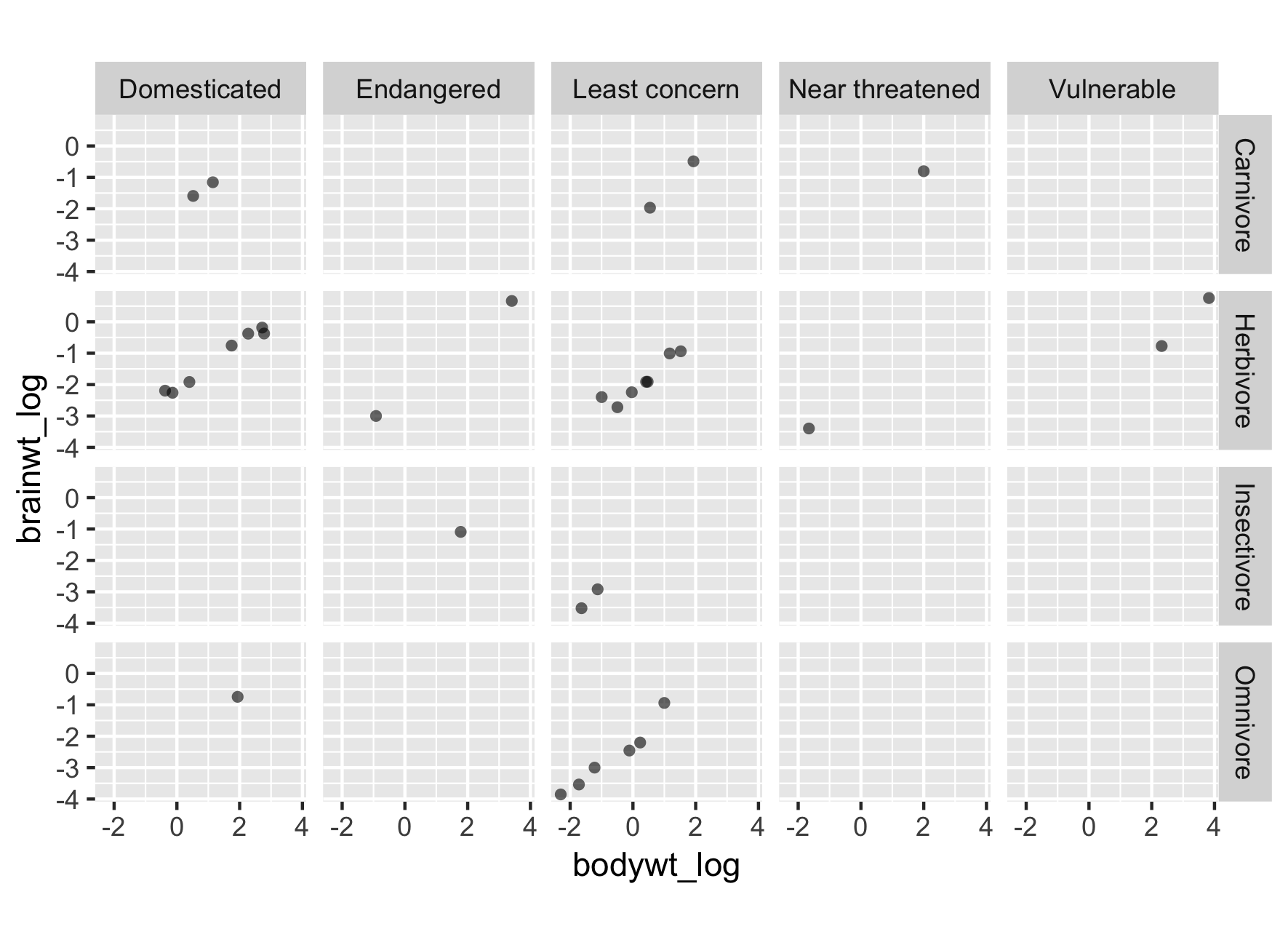
Reinitialize plot with new labels

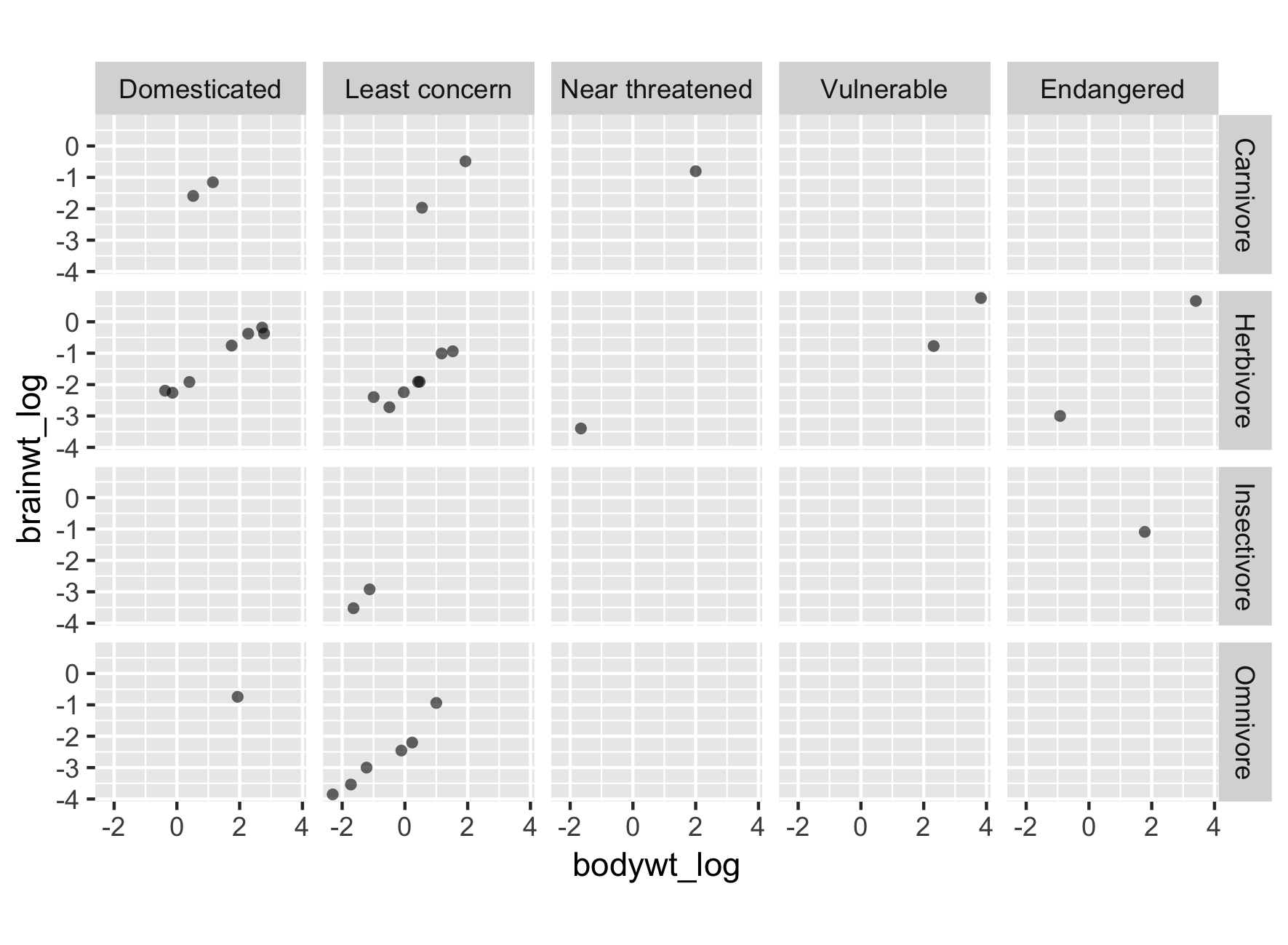
Changing the order of levels
# Change order of levels:
msleep2$conservation = fct_relevel(msleep2$conservation,
c("Domesticated",
"Least concern",
"Near threatened",
"Vulnerable",
"Endangered"))
Reinitialize plot with new order
Let's practice!
Intermediate Data Visualization with ggplot2

