Stats outside geoms
Intermediate Data Visualization with ggplot2

Rick Scavetta
Founder, Scavetta Academy
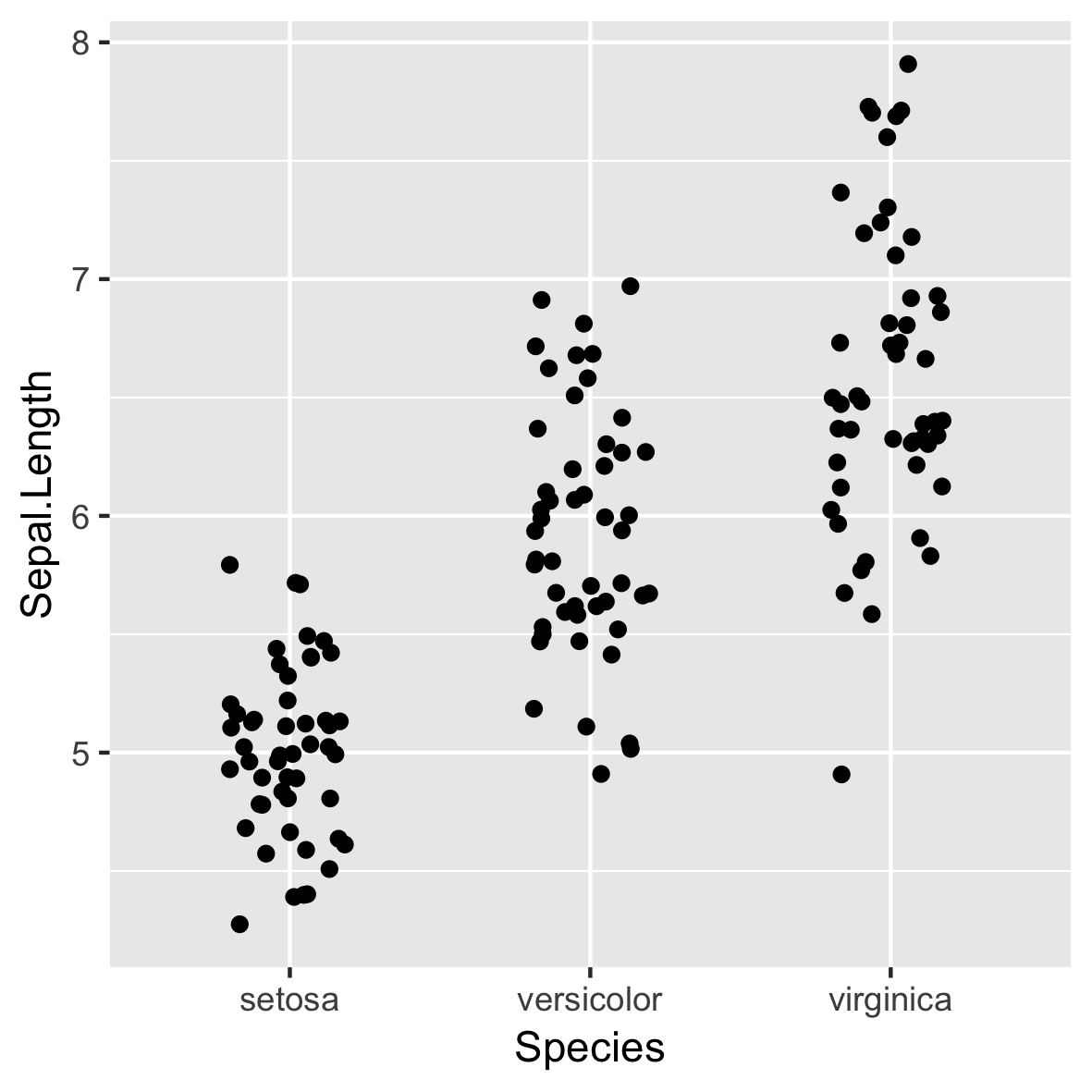
Basic plot
ggplot(iris, aes(x = Species,
y = Sepal.Length)) +
geom_jitter(width = 0.2)
Calculating statistics
set.seed(123)
xx <- rnorm(100)
mean(xx)
[1] 0.09040591
mean(xx) + (sd(xx) * c(-1, 1))
[1] -0.822410 1.003222
Calculating statistics
set.seed(123)
xx <- rnorm(100)
# Hmisc
library(Hmisc)
smean.sdl(xx, mult = 1)
Mean Lower Upper
0.09040591 -0.82240997 1.00322179
# ggplot2
mean_sdl(xx, mult = 1)
y ymin ymax
1 0.09040591 -0.82241 1.003222
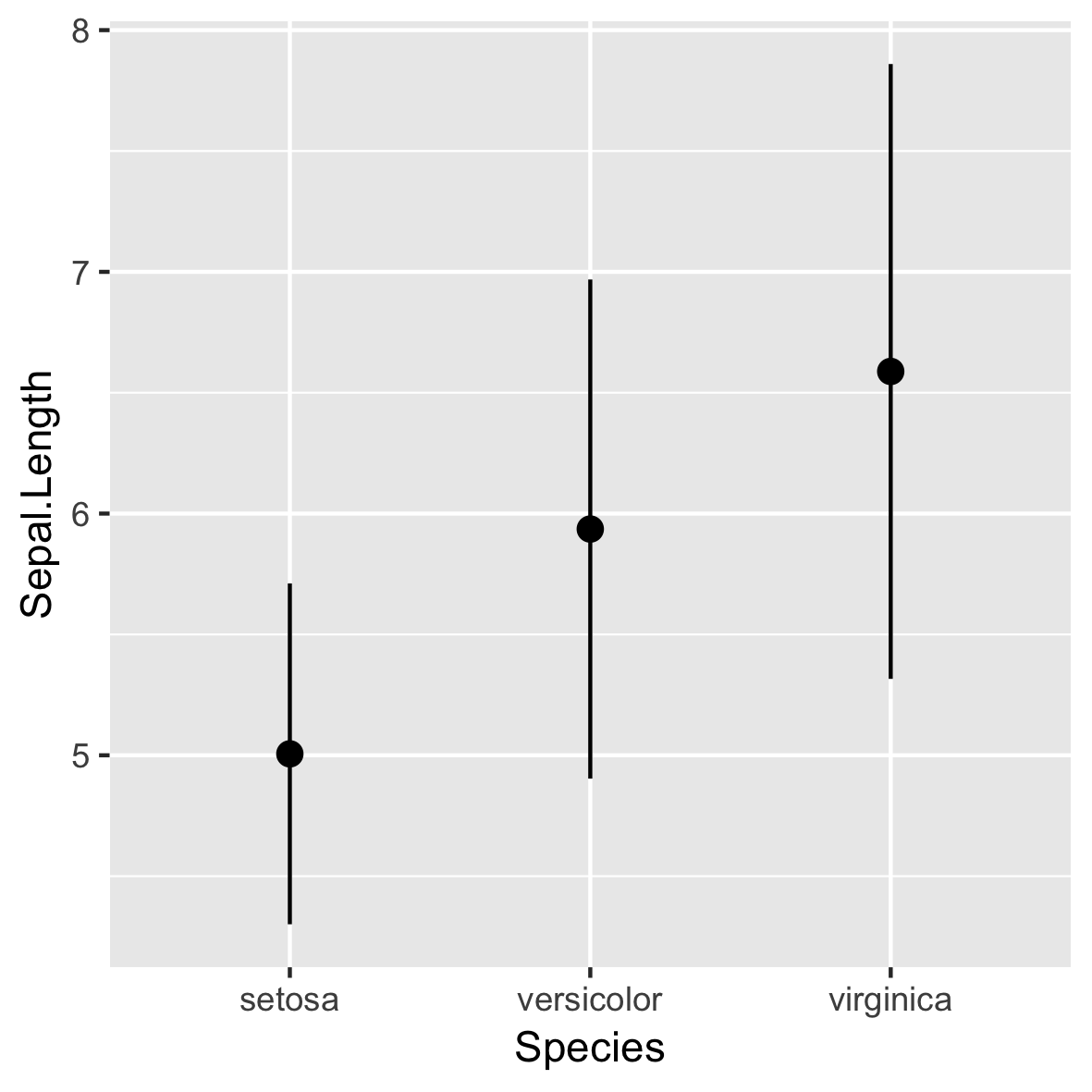
stat_summary()
ggplot(iris, aes(x = Species,
y = Sepal.Length)) +
stat_summary(fun.data = mean_sdl,
fun.args = list(mult = 1))
- Uses
geom_pointrange()by default
stat_summary()
ggplot(iris, aes(x = Species,
y = Sepal.Length)) +
stat_summary(fun = mean,
geom = "point") +
stat_summary(fun.data = mean_sdl,
fun.args = list(mult = 1),
geom = "errorbar",
width = 0.1)
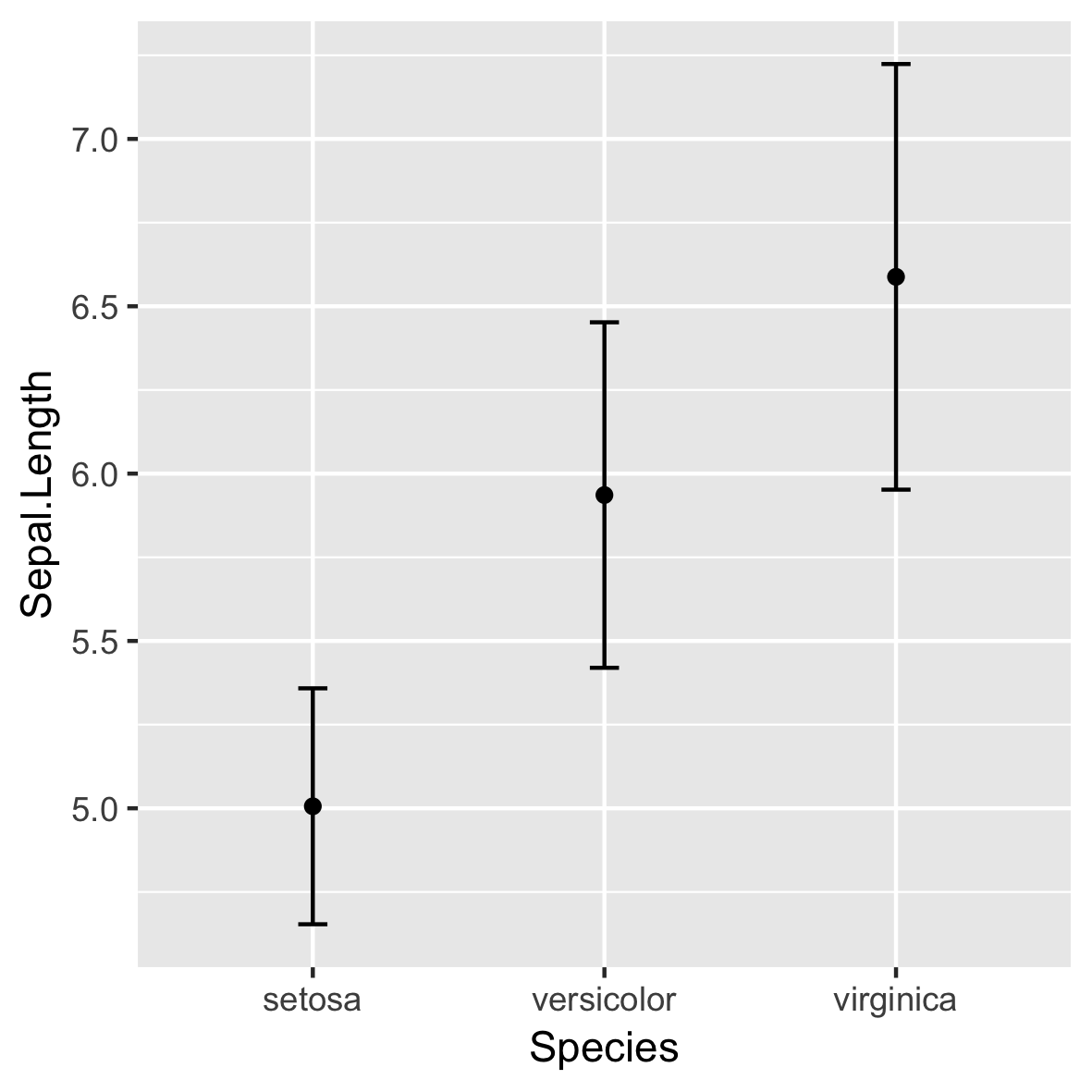
Not recommended!
95% confidence interval
ERR <- qt(0.975, length(xx) - 1) * (sd(xx) / sqrt(length(xx)))
mean(xx)
0.09040591
mean(xx) + (ERR * c(-1, 1)) # 95% CI
-0.09071657 0.27152838
mean_cl_normal(xx)
y ymin ymax
0.09040591 -0.09071657 0.2715284
Other stat_ functions
stat_ |
Description |
|---|---|
stat_summary() |
summarize y values at distinct x values. |
stat_function() |
compute y values from a function of x values. |
stat_qq() |
perform calculations for a quantile-quantile plot. |
MASS::mammals
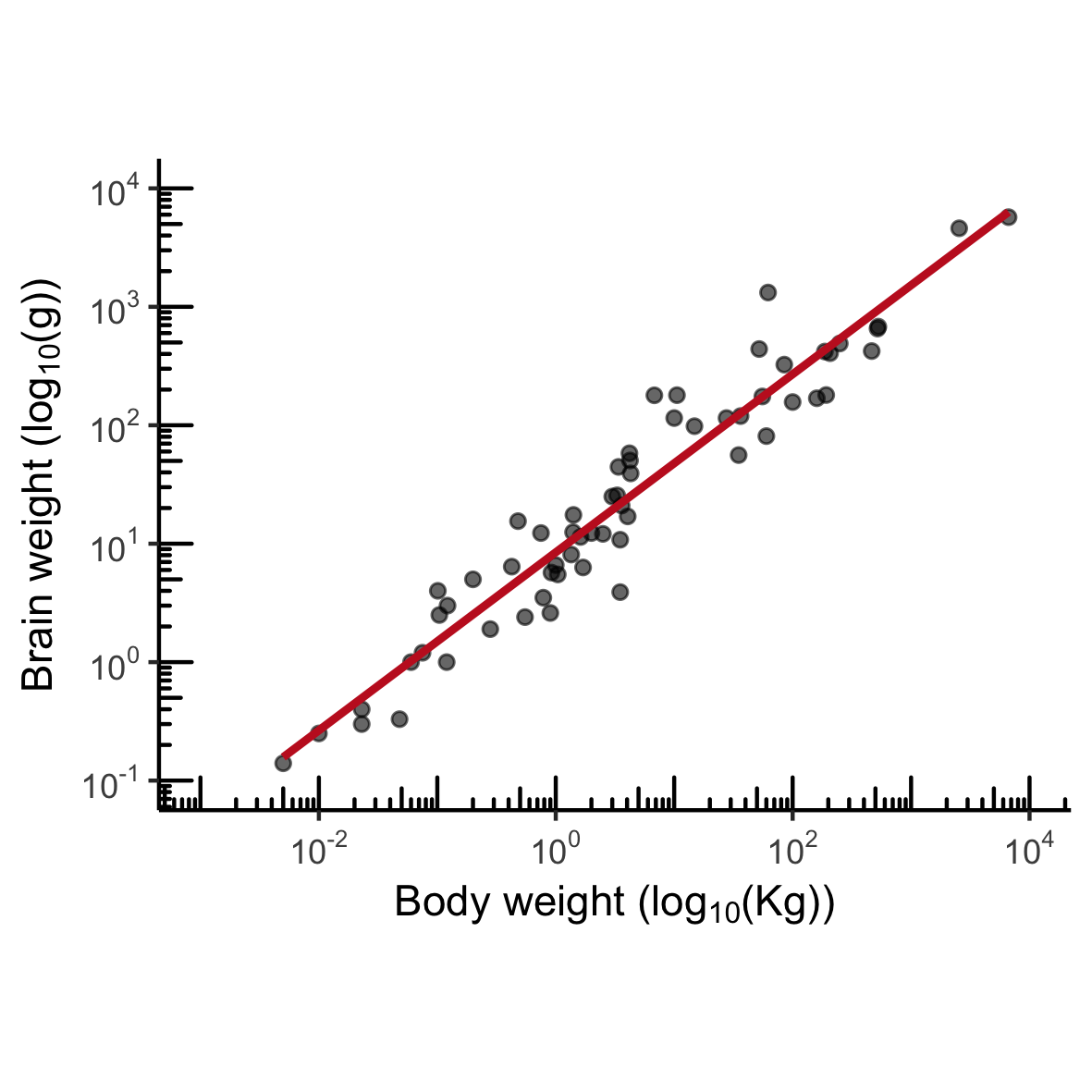
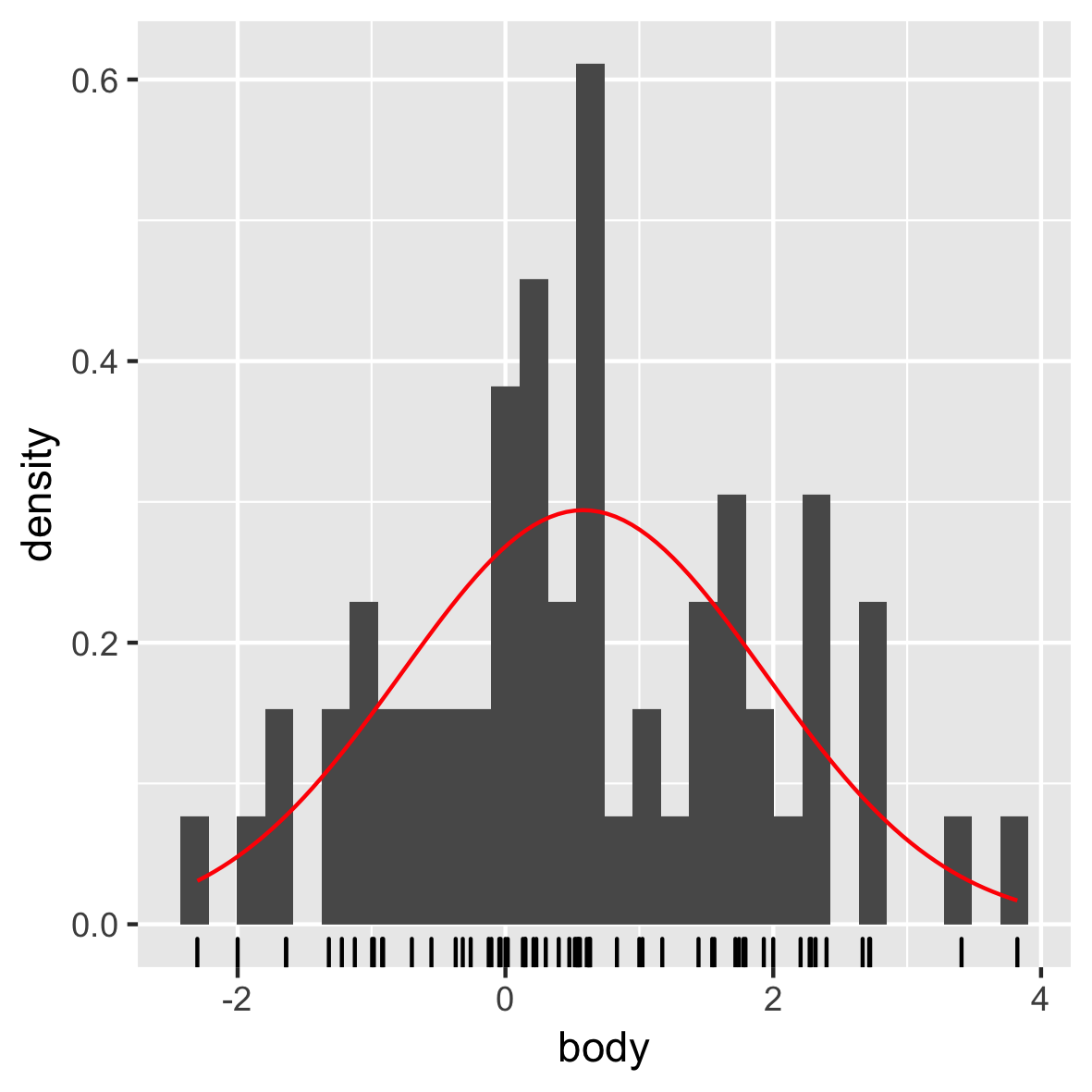
Normal distribution
mam.new <- data.frame(body = log10(mammals$body))
ggplot(mam.new, aes(x = body)) +
geom_histogram(aes( y = ..density..)) +
geom_rug() +
stat_function(fun = dnorm, color = "red",
args = list(mean = mean(mam.new$body),
sd = sd(mam.new$body)))
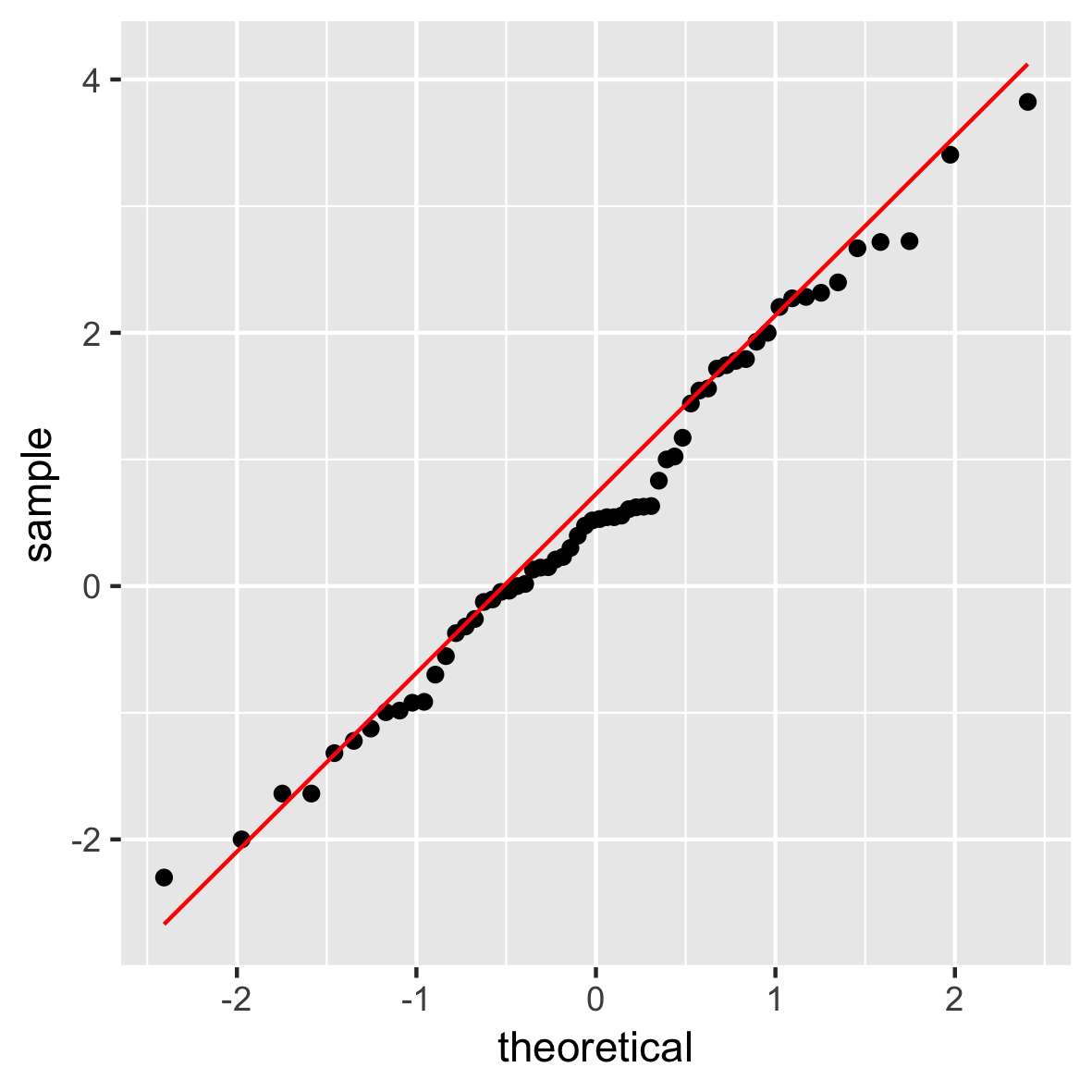
QQ plot
ggplot(mam.new, aes(sample = body)) +
stat_qq() +
geom_qq_line(col = "red")
Your turn!
Intermediate Data Visualization with ggplot2

