Evaluating a clustering
Unsupervised Learning in Python

Benjamin Wilson
Director of Research at lateral.io
Evaluating a clustering
- Can check correspondence with e.g. iris species
- ... but what if there are no species to check against?
- Measure quality of a clustering
- Informs choice of how many clusters to look for
Iris: clusters vs species
- k-means found 3 clusters amongst the iris samples
- Do the clusters correspond to the species?
species setosa versicolor virginica
labels
0 0 2 36
1 50 0 0
2 0 48 14
Cross tabulation with pandas
- Clusters vs species is a "cross-tabulation"
- Use the
pandaslibrary - Given the species of each sample as a list
species
print(species)
['setosa', 'setosa', 'versicolor', 'virginica', ... ]
Aligning labels and species
import pandas as pd
df = pd.DataFrame({'labels': labels, 'species': species})
print(df)
labels species
0 1 setosa
1 1 setosa
2 2 versicolor
3 2 virginica
4 1 setosa
...
Crosstab of labels and species
ct = pd.crosstab(df['labels'], df['species'])
print(ct)
species setosa versicolor virginica
labels
0 0 2 36
1 50 0 0
2 0 48 14
How to evaluate a clustering, if there were no species information?
Measuring clustering quality
Using only samples and their cluster labels
A good clustering has tight clusters
Samples in each cluster bunched together
Inertia measures clustering quality
- Measures how spread out the clusters are (lower is better)
- Distance from each sample to centroid of its cluster
- After
fit(), available as attributeinertia_ - k-means attempts to minimize the inertia when choosing clusters
from sklearn.cluster import KMeans
model = KMeans(n_clusters=3)
model.fit(samples)
print(model.inertia_)
78.9408414261
The number of clusters
- Clusterings of the iris dataset with different numbers of clusters
- More clusters means lower inertia
- What is the best number of clusters?
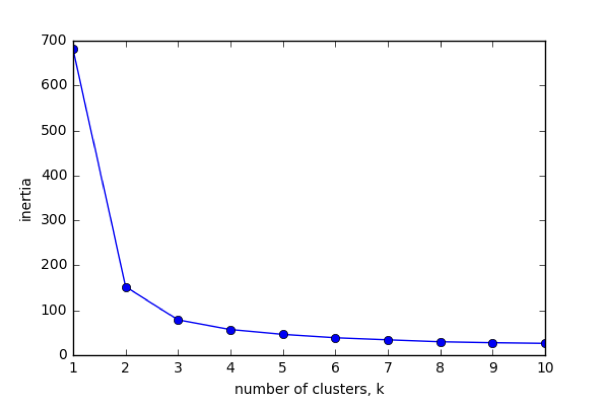
How many clusters to choose?
- A good clustering has tight clusters (so low inertia)
- ... but not too many clusters!
- Choose an "elbow" in the inertia plot
- Where inertia begins to decrease more slowly
- E.g., for iris dataset, 3 is a good choice

Let's practice!
Unsupervised Learning in Python

