Quantifying model fit
Introduction to Regression in R

Richie Cotton
Data Evangelist at DataCamp
Bream and perch models
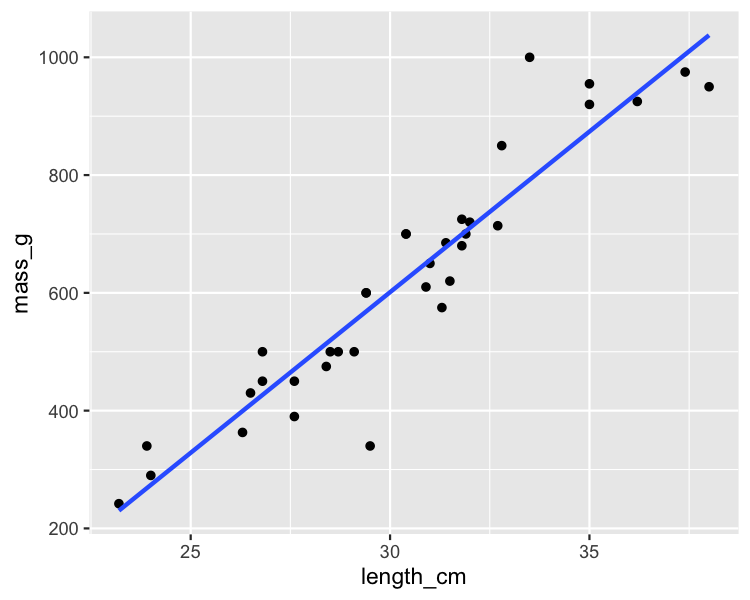
Bream
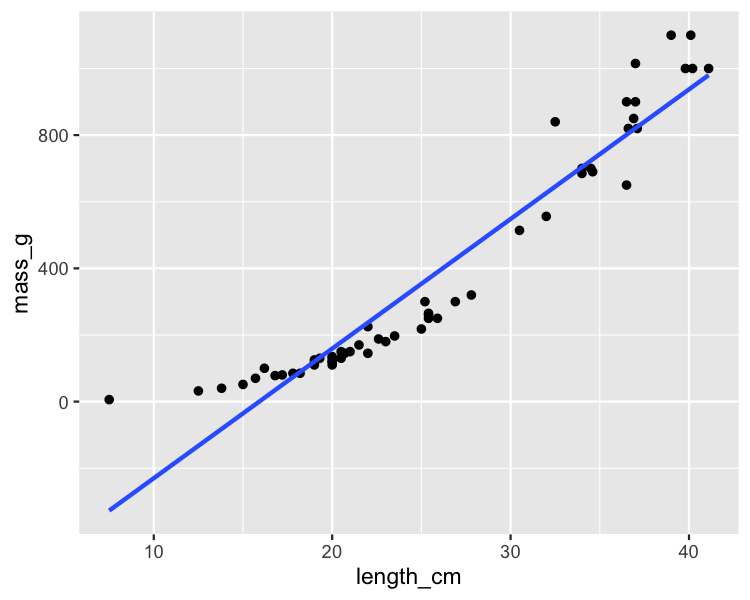
Perch
Coefficient of determination
Sometimes called "r-squared" or "R-squared".
the proportion of the variance in the response variable that is predictable from the explanatory variable
1means a perfect fit0means the worst possible fit
summary()
Look at the value titled "Multiple R-Squared"
mdl_bream <- lm(mass_g ~ length_cm, data = bream)
summary(mdl_bream)
# Some lines of output omitted
Residual standard error: 74.15 on 33 degrees of freedom
Multiple R-squared: 0.8781, Adjusted R-squared: 0.8744
F-statistic: 237.6 on 1 and 33 DF, p-value: < 2.2e-16
glance()
library(broom)
library(dplyr)
mdl_bream %>%
glance()
# A tibble: 1 × 12
r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 0.878 0.874 74.2 238. 1.22e-16 1 -199. 405. 409.
# ... with 3 more variables: deviance <dbl>, df.residual <int>, nobs <int>
mdl_bream %>%
glance() %>%
pull(r.squared)
0.8780627
It's just correlation squared
bream %>%
summarize(
coeff_determination = cor(length_cm, mass_g) ^ 2
)
coeff_determination
1 0.8780627
Residual standard error (RSE)
a "typical" difference between a prediction and an observed response
It has the same unit as the response variable.
summary() again
Look at the value titled "Residual standard error"
summary(mdl_bream)
# Some lines of output omitted
Residual standard error: 74.15 on 33 degrees of freedom
Multiple R-squared: 0.8781, Adjusted R-squared: 0.8744
F-statistic: 237.6 on 1 and 33 DF, p-value: < 2.2e-16
glance() again
library(broom)
library(dplyr)
mdl_bream %>%
glance()
# A tibble: 1 x 11
r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC deviance df.residual
<dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <int>
1 0.878 0.874 74.2 238. 1.22e-16 2 -199. 405. 409. 181452. 33
mdl_bream %>%
glance() %>%
pull(sigma)
74.15224
Calculating RSE: residuals squared
bream %>%
mutate(
residuals_sq = residuals(mdl_bream) ^ 2
)
species mass_g length_cm residuals_sq
1 Bream 242 23.2 138.9571
2 Bream 290 24.0 260.7586
3 Bream 340 23.9 5126.9926
4 Bream 363 26.3 1318.9197
5 Bream 430 26.5 390.9743
6 Bream 450 26.8 547.9380
...
Calculating RSE: sum of residuals squared
bream %>%
mutate(
residuals_sq = residuals(mdl_bream) ^ 2
) %>%
summarize(
resid_sum_of_sq = sum(residuals_sq)
)
resid_sum_of_sq
1 181452.3
Calculating RSE: degrees of freedom
Degrees of freedom equals the number of observations minus the number of model coefficients.
bream %>%
mutate(
residuals_sq = residuals(mdl_bream) ^ 2
) %>%
summarize(
resid_sum_of_sq = sum(residuals_sq),
deg_freedom = n() - 2
)
resid_sum_of_sq deg_freedom
1 181452.3 33
Calculating RSE: square root of ratio
bream %>%
mutate(
residuals_sq = residuals(mdl_bream) ^ 2
) %>%
summarize(
resid_sum_of_sq = sum(residuals_sq),
deg_freedom = n() - 2,
rse = sqrt(resid_sum_of_sq / deg_freedom)
)
resid_sum_of_sq deg_freedom rse
1 181452.3 33 74.15224
Interpreting RSE
mdl_bream has an RSE of 74.
The difference between predicted bream masses and observed bream masses is typically about 74g.
Root-mean-square error (RMSE)
Residual standard error
bream %>%
mutate(
residuals_sq = residuals(mdl_bream) ^ 2
) %>%
summarize(
resid_sum_of_sq = sum(residuals_sq),
deg_freedom = n() - 2,
rse = sqrt(resid_sum_of_sq / deg_freedom)
)
Root-mean-square error
bream %>%
mutate(
residuals_sq = residuals(mdl_bream) ^ 2
) %>%
summarize(
resid_sum_of_sq = sum(residuals_sq),
n_obs = n(),
rmse = sqrt(resid_sum_of_sq / n_obs)
)
Let's practice!
Introduction to Regression in R

