Maße der zentralen Tendenz
Einführung in die Statistik in Python

Maggie Matsui
Content Developer, DataCamp
Daten zum Schlaf von Säugetieren
print(msleep)
name genus vore order ... sleep_cycle awake brainwt bodywt
1 Cheetah Acinonyx carni Carnivora ... NaN 11.9 NaN 50.000
2 Owl monkey Aotus omni Primates ... NaN 7.0 0.01550 0.480
3 Mountain beaver Aplodontia herbi Rodentia ... NaN 9.6 NaN 1.350
4 Greater short-ta... Blarina omni Soricomorpha ... 0.133333 9.1 0.00029 0.019
5 Cow Bos herbi Artiodactyla ... 0.666667 20.0 0.42300 600.000
.. ... ... ... ... ... ... ... ... ...
79 Tree shrew Tupaia omni Scandentia ... 0.233333 15.1 0.00250 0.104
80 Bottle-nosed do... Tursiops carni Cetacea ... NaN 18.8 NaN 173.330
81 Genet Genetta carni Carnivora ... NaN 17.7 0.01750 2.000
82 Arctic fox Vulpes carni Carnivora ... NaN 11.5 0.04450 3.380
83 Red fox Vulpes carni Carnivora ... 0.350000 14.2 0.05040 4.230
Histogramme
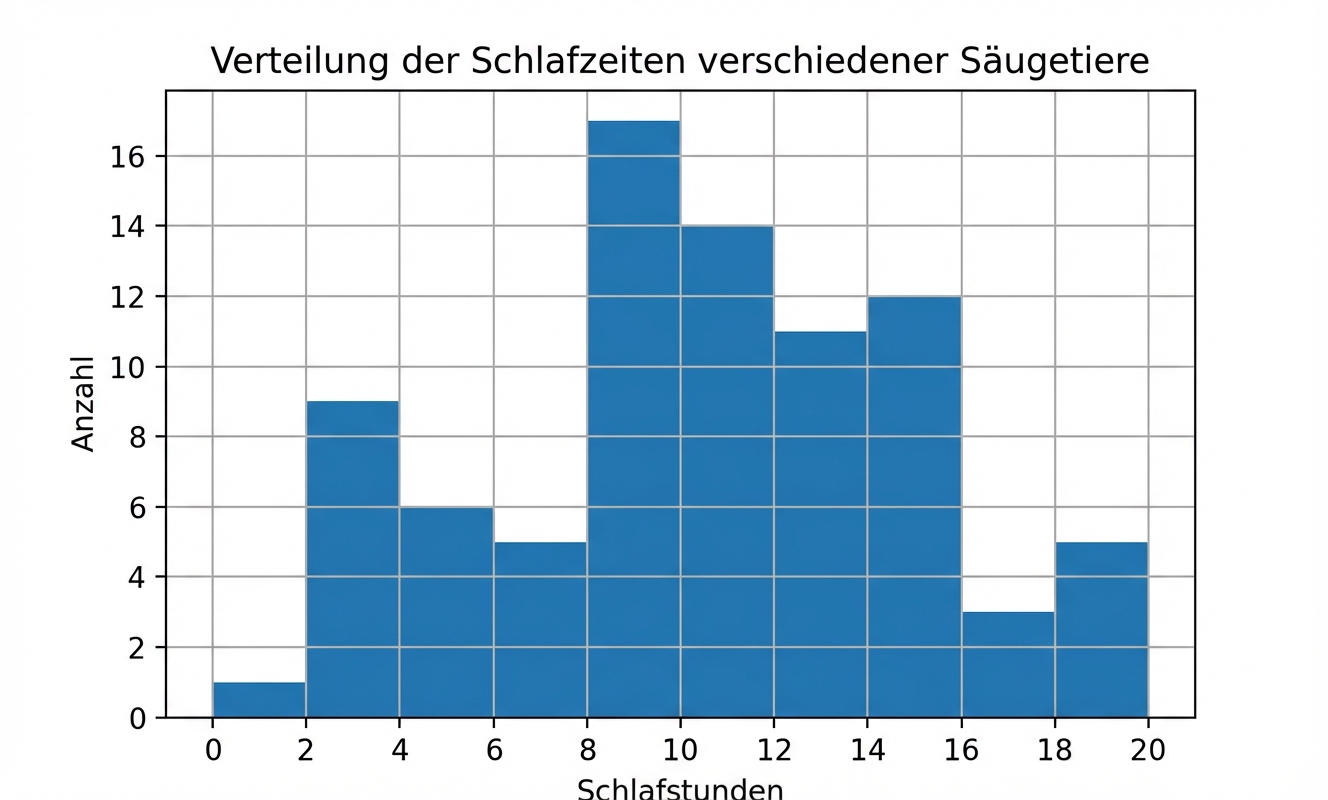
Wie lange schlafen die Säugetiere in diesem Datensatz normalerweise?
Was ist ein typischer Wert?
Wo liegt der Mittelpunkt der Daten?
- Mittelwert
- Median
- Modus
Maße der zentralen Tendenz: Mittelwert
name sleep_total
1 Cheetah 12.1
2 Owl monkey 17.0
3 Mountain beaver 14.4
4 Greater short-t... 14.9
5 Cow 4.0
.. ... ...
$\text{Mean sleep time}=$
$$\frac{12.1 + 17.0 + 14.4 + 14.9 + ...}{83} = 10.43$$
import numpy as np
np.mean(msleep['sleep_total'])
10.43373
Maße der zentralen Tendenz: Median
msleep['sleep_total'].sort_values()
29 1.9
30 2.7
22 2.9
9 3.0
23 3.1
...
19 18.0
61 18.1
36 19.4
21 19.7
42 19.9
msleep['sleep_total'].sort_values().iloc[41]
10.1
np.median(msleep['sleep_total'])
10.1
Maße der zentralen Tendenz: Modus
Häufigster Wert
msleep['sleep_total'].value_counts()
12.5 4
10.1 3
14.9 2
11.0 2
8.4 2
...
14.3 1
17.0 1
Name: sleep_total, Length: 65, dtype: int64
msleep['vore'].value_counts()
herbi 32
omni 20
carni 19
insecti 5
Name: vore, dtype: int64
import statistics
statistics.mode(msleep['vore'])
'herbi'
Einen Ausreißer hinzufügen
# Subset msleep to select rows where 'vore' equals 'insecti'
msleep[msleep['vore'] == 'insecti']
name genus vore order sleep_total
22 Big brown bat Eptesicus insecti Chiroptera 19.7
43 Little brown bat Myotis insecti Chiroptera 19.9
62 Giant armadillo Priodontes insecti Cingulata 18.1
67 Eastern american mole Scalopus insecti Soricomorpha 8.4
Einen Ausreißer hinzufügen
msleep[msleep['vore'] == "insecti"]['sleep_total'].agg([np.mean, np.median])
mean 16.53
median 18.9
Name: sleep_total, dtype: float64
Einen Ausreißer hinzufügen
msleep[msleep['vore'] == 'insecti']
name genus vore order sleep_total 22 Big brown bat Eptesicus insecti Chiroptera 19.7 43 Little brown bat Myotis insecti Chiroptera 19.9 62 Giant armadillo Priodontes insecti Cingulata 18.1 67 Eastern american mole Scalopus insecti Soricomorpha 8.484 Mystery insectivore ... insecti ... 0.0
Einen Ausreißer hinzufügen
msleep[msleep['vore'] == "insecti"]['sleep_total'].agg([np.mean, np.median])
mean 13.22
median 18.1
Name: sleep_total, dtype: float64
Mittelwert: 16,5 → 13,2
Median: 18,9 → 18,1
Welches Maß sollten wir verwenden
# Import matplotlib.pyplot with alias plt
import matplotlib.pyplot as plt
# Histogram of values
data['values'].hist()
# Show the plot
plt.show()
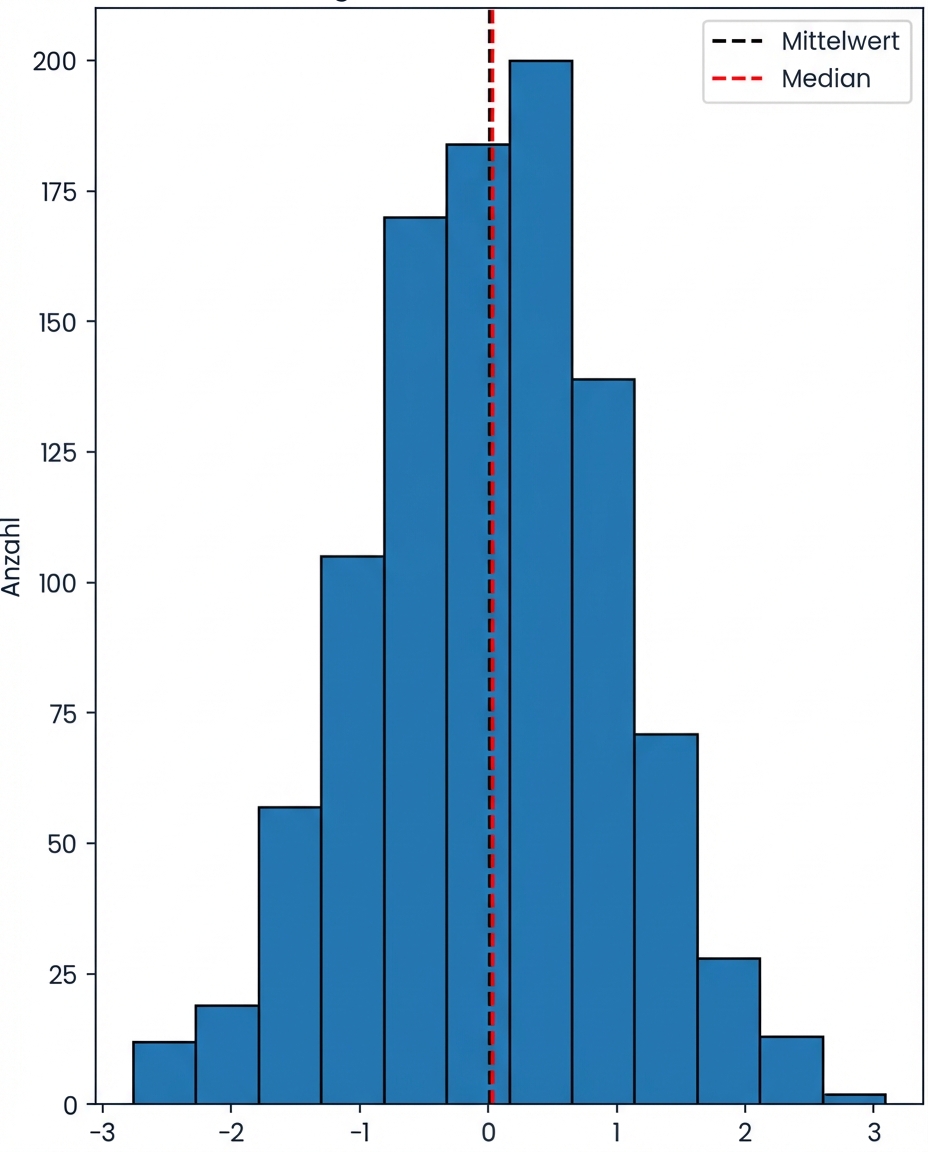
Schieflage
Linksschief
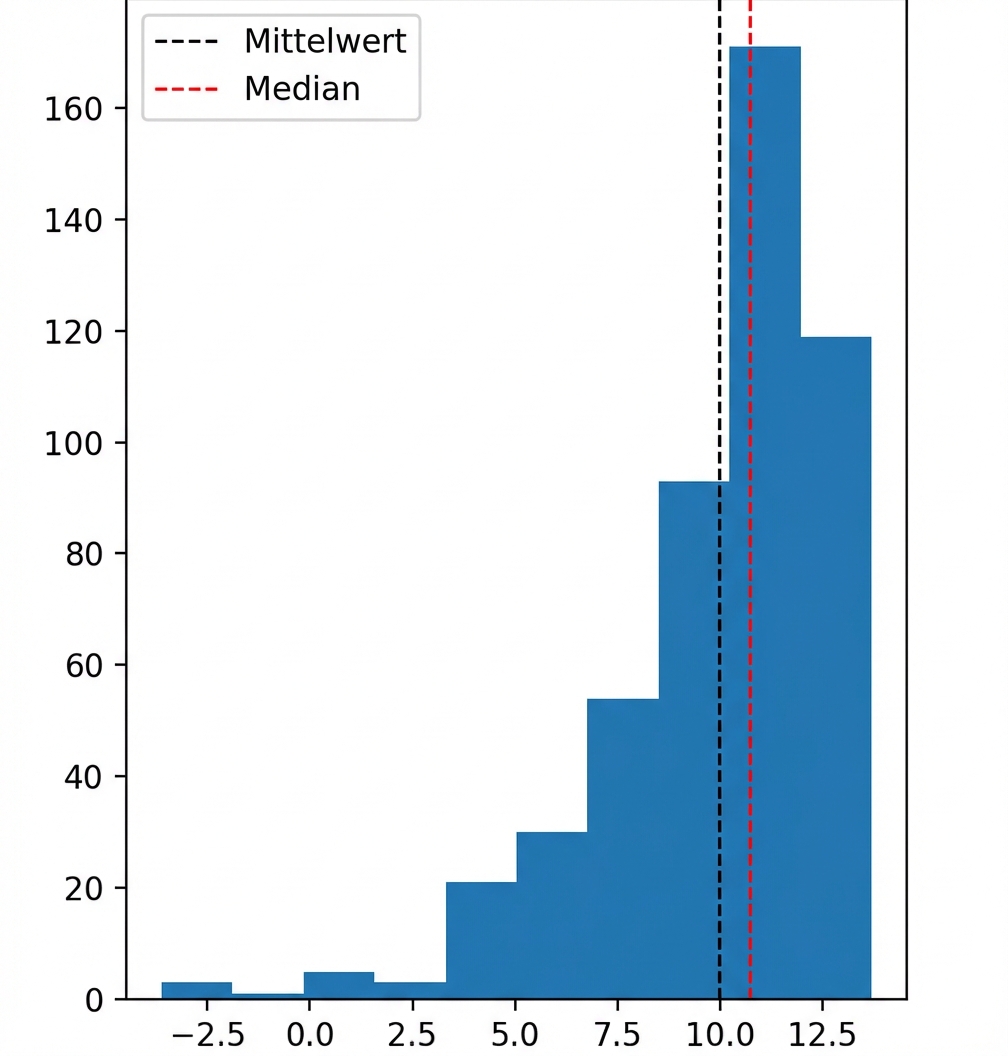
Rechtsschief
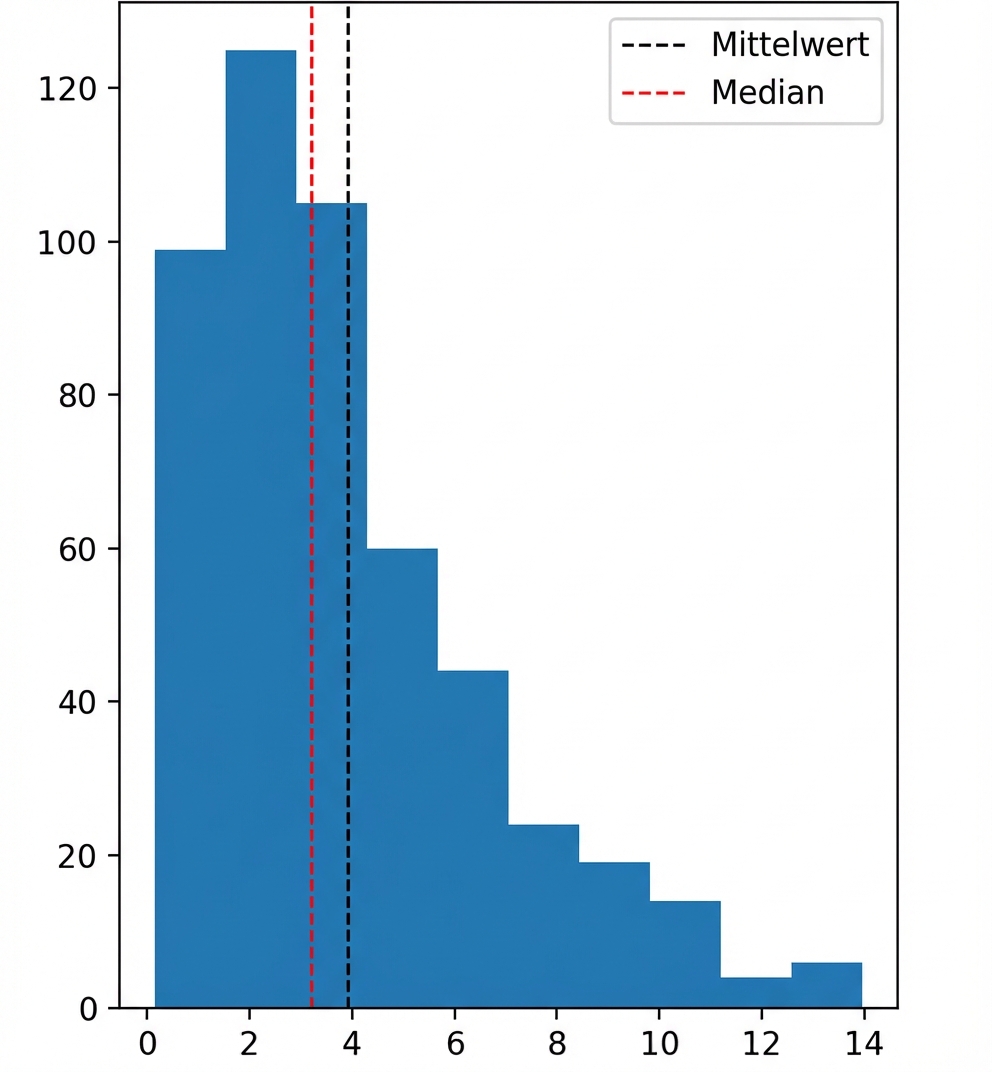
Welches Maß sollten wir verwenden


Lass uns üben!
Einführung in die Statistik in Python

