Maße der zentralen Tendenz
Einführung in die Statistik in R

Maggie Matsui
Content Developer, DataCamp
Daten zum Schlaf von Säugetieren
msleep
# A tibble: 83 x 11
name genus vore order sleep_total sleep_rem sleep_cycle awake
<chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Cheetah Acinonyx carni Carnivora 12.1 NA NA 11.9
2 Owl monkey Aotus omni Primates 17 1.8 NA 7
3 Mountain beaver Aplodontia herbi Rodentia 14.4 2.4 NA 9.6
4 Greater short... Blarina omni Soricomorpha 14.9 2.3 0.133 9.1
5 Cow Bos herbi Artiodactyla 4 0.7 0.667 20
6 Three-toed sloth Bradypus herbi Pilosa 14.4 2.2 0.767 9.6
7 Northern fur... Callorhinus carni Carnivora 8.7 1.4 0.383 15.3
# ... with 76 more rows, and 2 more variables: brainwt <dbl>, bodywt <dbl>
Histogramme
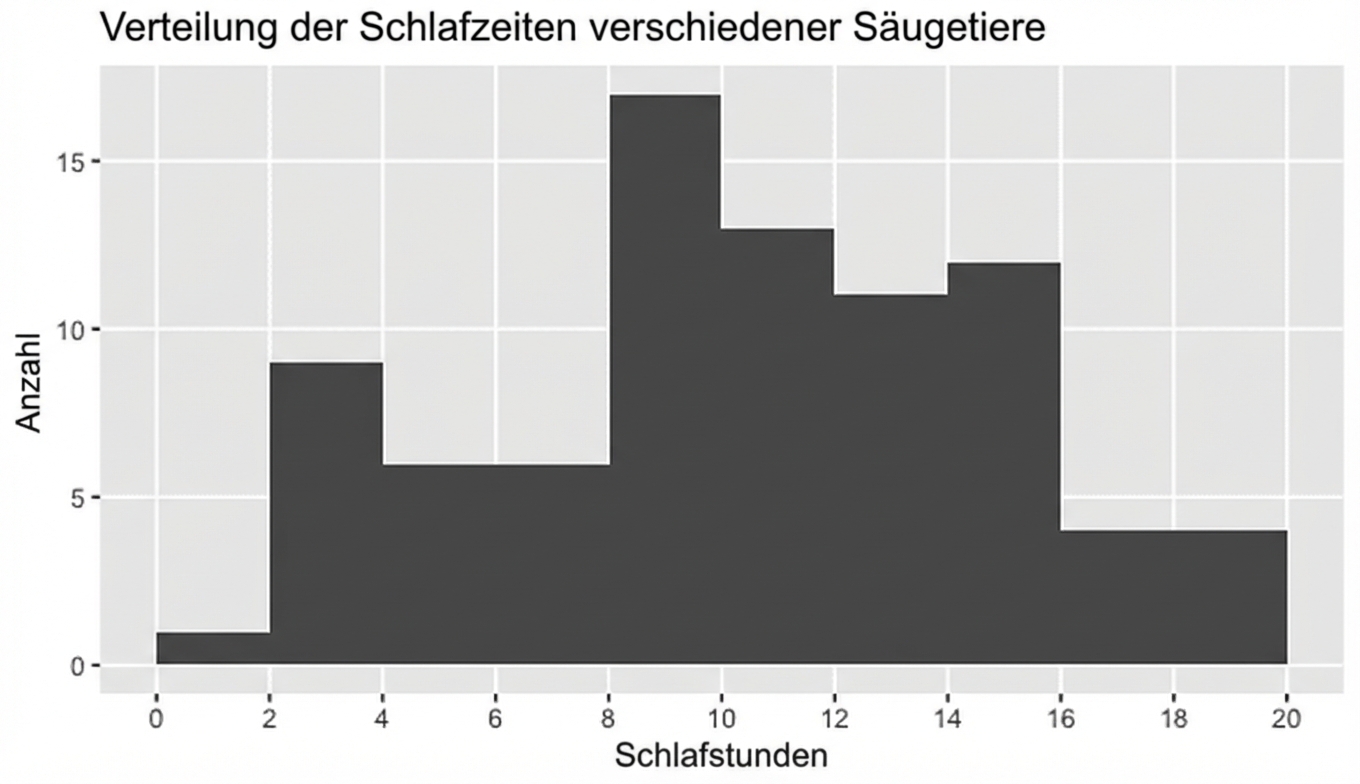
Wie lange schlafen die Säugetiere in diesem Datensatz normalerweise?
Was ist ein typischer Wert?
Wo liegt der Mittelpunkt der Daten?
- Mittelwert
- Median
- Modus
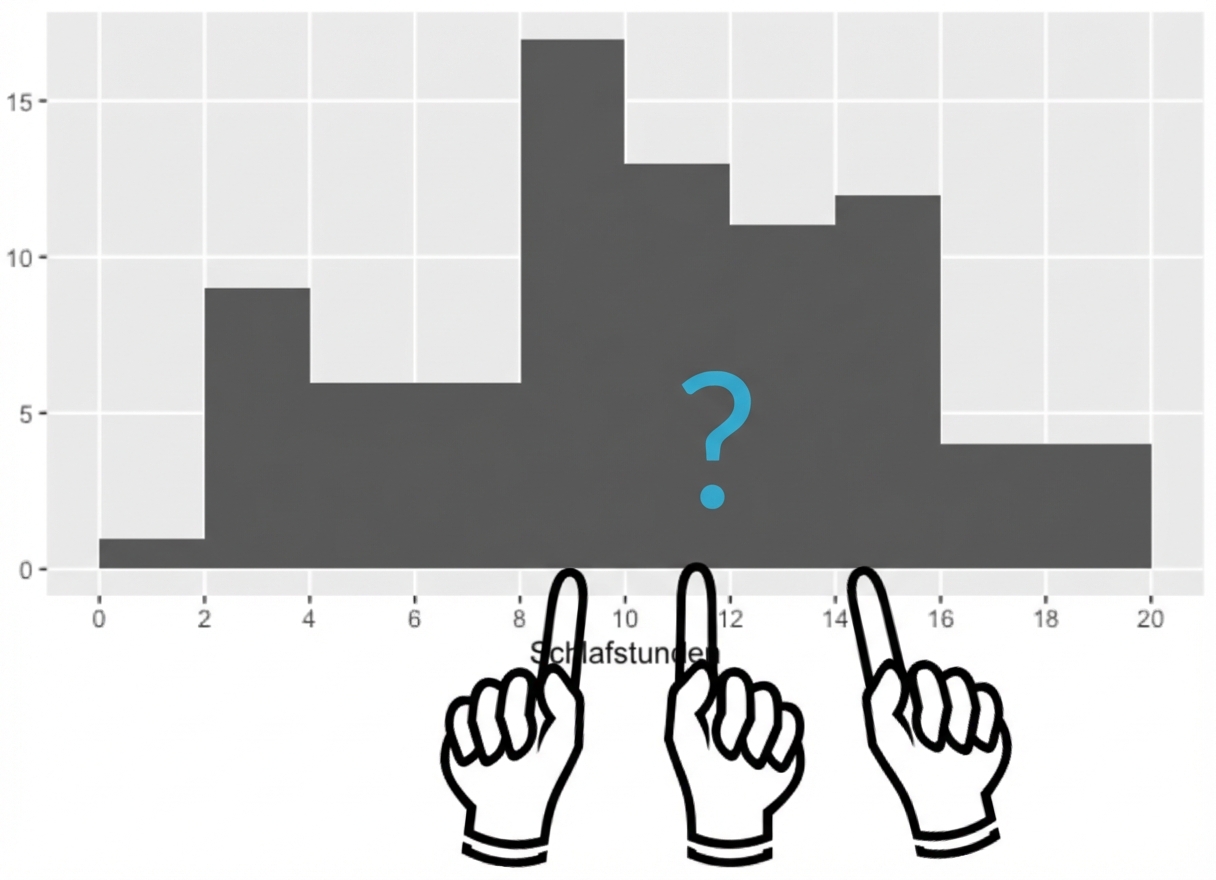
Maße der zentralen Tendenz: Mittelwert
name sleep_total
1 Cheetah 12.1
2 Owl monkey 17.0
3 Mountain beaver 14.4
4 Greater short-tailed shrew 14.9
...
$$\text{Durschnittliche Schlafzeit}=\frac{12.1 + 17.0 + 14.4 + 14.9 + ...}{83} = 10.43$$
mean(msleep$sleep_total)
10.43373
Maße der zentralen Tendenz: Median
sort(msleep$sleep_total)
[1] 1.9 2.7 2.9 3.0 3.1 3.3 3.5 3.8 3.9 4.0 4.4 5.2 5.3 5.3 5.4 5.6 6.2
...
[52] 11.5 12.1 12.5 12.5 12.5 12.5 12.8 12.8 13.0 13.5 13.7 13.8 14.2 14.3 14.4 14.4 14.5
[69] 14.6 14.9 14.9 15.6 15.8 15.8 15.9 16.6 17.0 17.4 18.0 18.1 19.4 19.7 19.9
sort(msleep$sleep_total)[42]
10.1
median(msleep$sleep_total)
10.1
Maße der zentralen Tendenz: Modus
Häufigster Wert
# Count and sort 'sleep_total' descending
msleep %>% count(sleep_total, sort = TRUE)
sleep_total n
<dbl> <int>
1 12.5 4
2 10.1 3
3 5.3 2
4 6.3 2
...
# Count and sort 'vore' descending
msleep %>% count(vore, sort = TRUE)
vore n
<chr> <int>
1 herbi 32
2 omni 20
3 carni 19
4 NA 7
5 insecti 5
Gefilterte Daten für Insektenfresser
msleep %>%
filter(vore == "insecti")
name genus vore order sleep_total
<chr> <chr> <chr> <chr> <dbl>
1 Big brown bat Eptesicus insecti Chiroptera 19.7
2 Little brown bat Myotis insecti Chiroptera 19.9
3 Giant armadillo Priodontes insecti Cingulata 18.1
4 Eastern american mole Scalopus insecti Soricomorpha 8.4
Zusammenfassung der Statistiken zum Schlaf bei Insektenfressern
msleep %>% filter(vore == "insecti") %>%summarize(mean_sleep = mean(sleep_total), median_sleep = median(sleep_total))
mean_sleep median_sleep
<dbl> <dbl>
1 16.52 18.9
Einen Ausreißer hinzufügen
msleep %>%
filter(vore == "insecti")
name genus vore order sleep_total
<chr> <chr> <chr> <chr> <dbl>
1 Big brown bat Eptesicus insecti Chiroptera 19.7
2 Little brown bat Myotis insecti Chiroptera 19.9
3 Giant armadillo Priodontes insecti Cingulata 18.1
4 Eastern american mole Scalopus insecti Soricomorpha 8.4
5 Mystery insectivore ... ... ... 0.0
Zusammenfassende Statistik nach dem Hinzufügen eines Ausreißers
msleep %>% filter(vore == "insecti") %>%summarize(mean_sleep = mean(sleep_total), median_sleep = median(sleep_total))
mean_sleep median_sleep
<dbl> <dbl>
1 13.22 18.1
Mittelwert: 16,5 → 13,2
Median: 18,9 → 18,1
Welches Maß sollten wir verwenden?
# Create histogram of insecti
msleep %>%
filter(vore == "insecti") %>%
ggplot(aes(insecti)) +
geom_histogram()
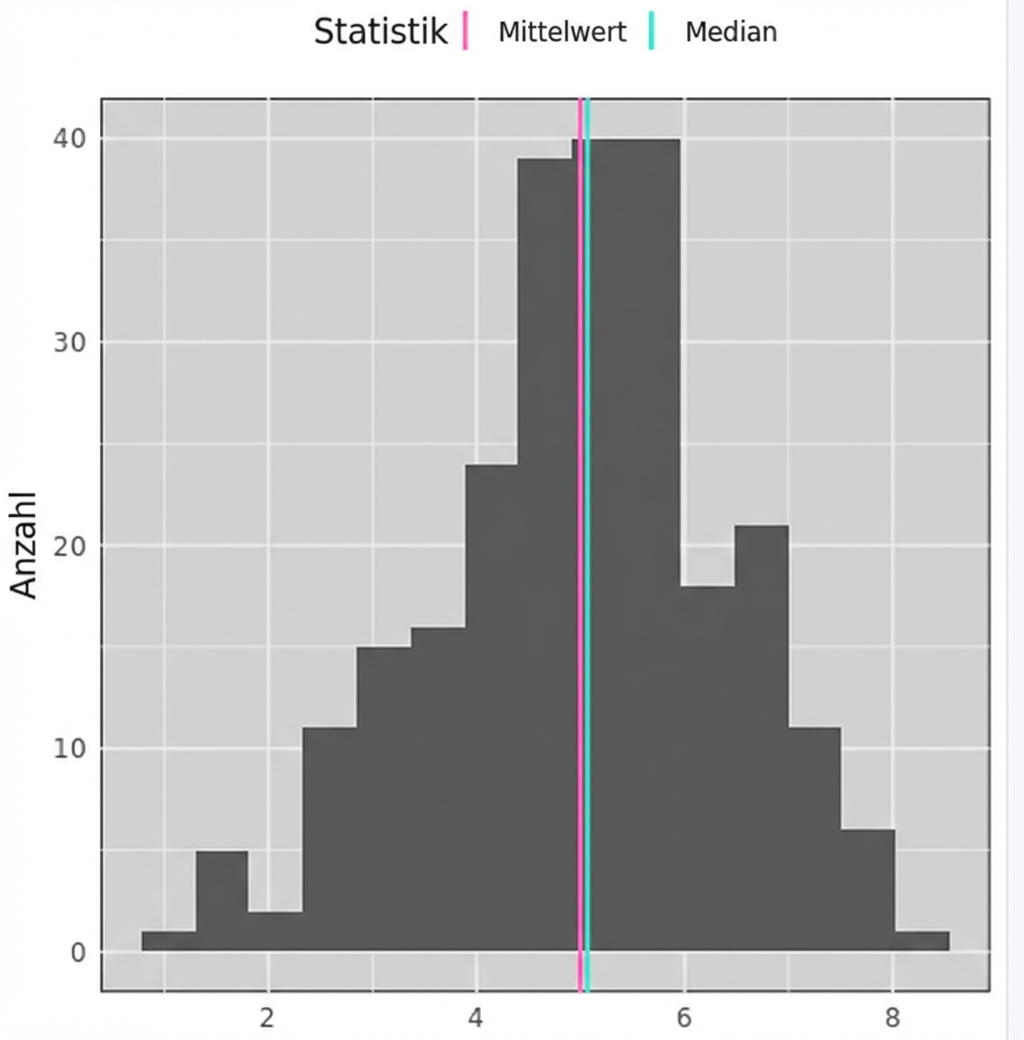
Schieflage
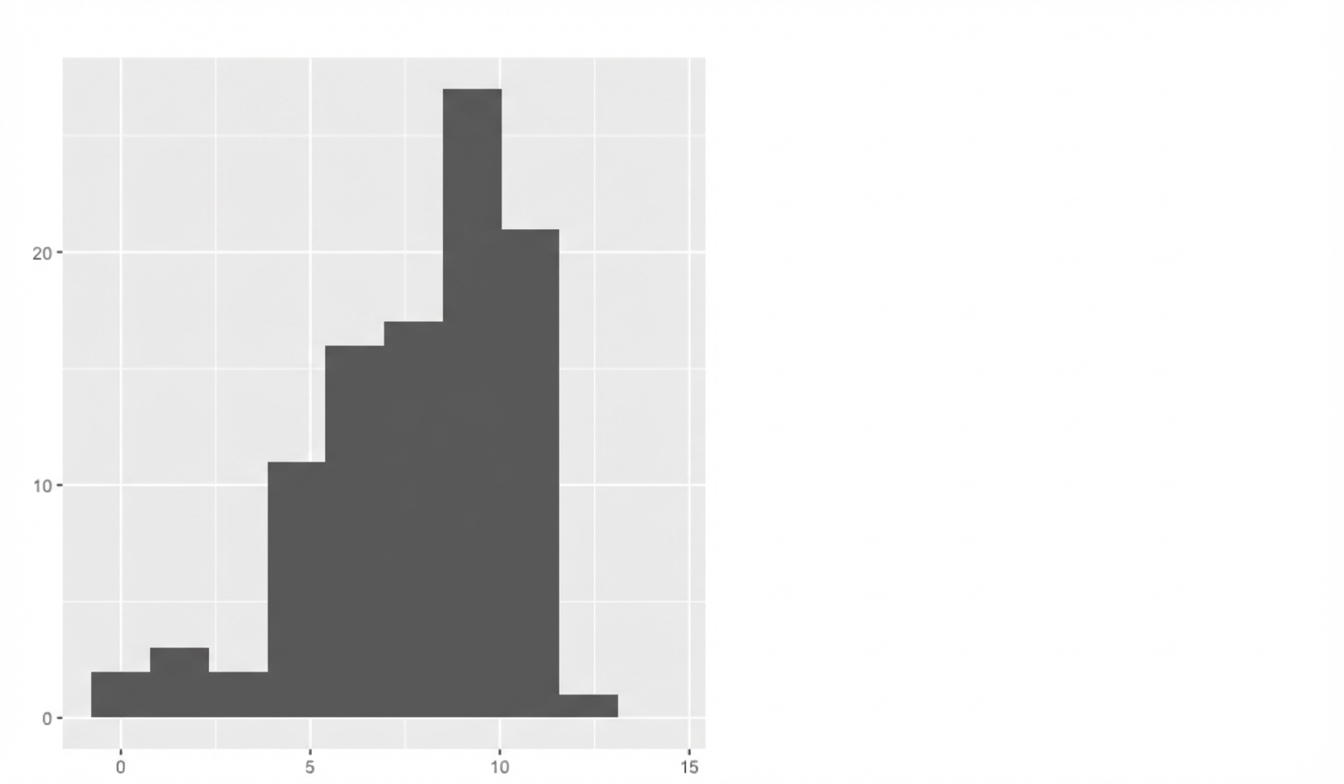
Schieflage
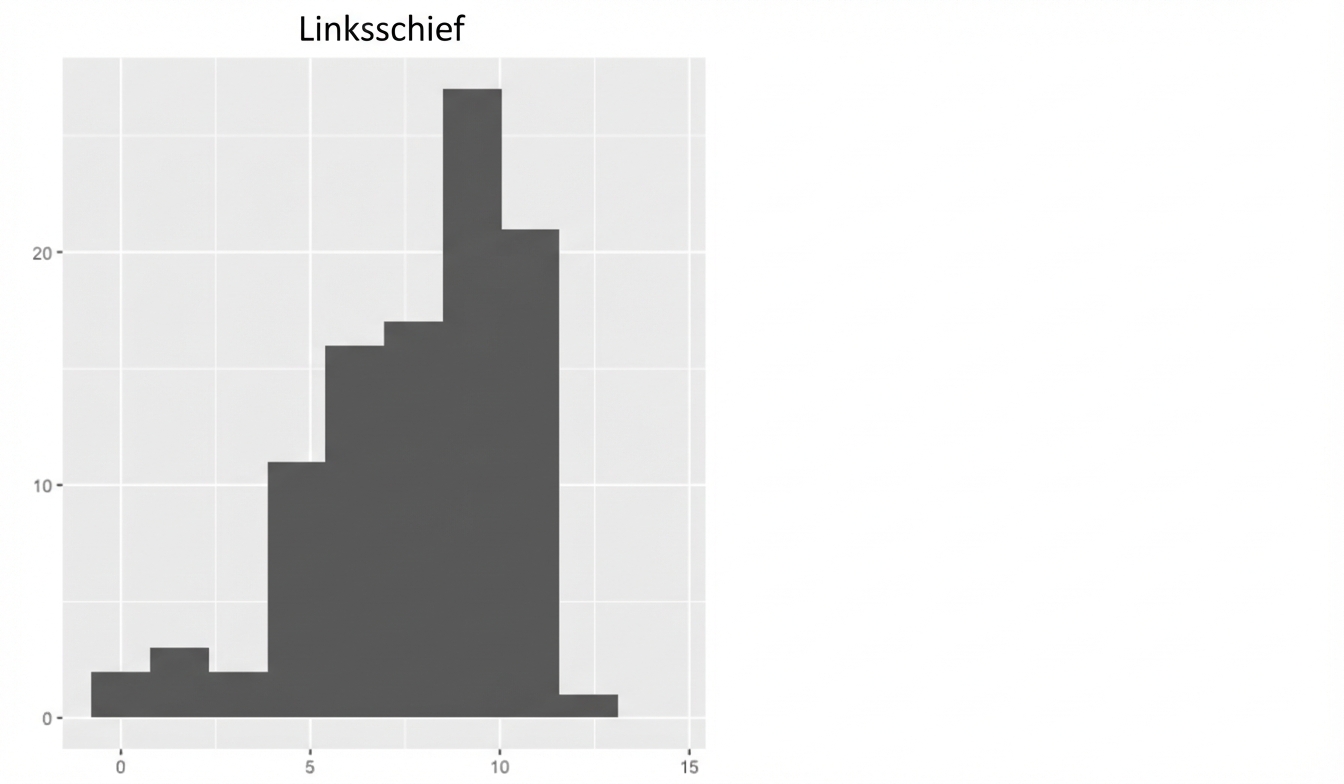
Schieflage
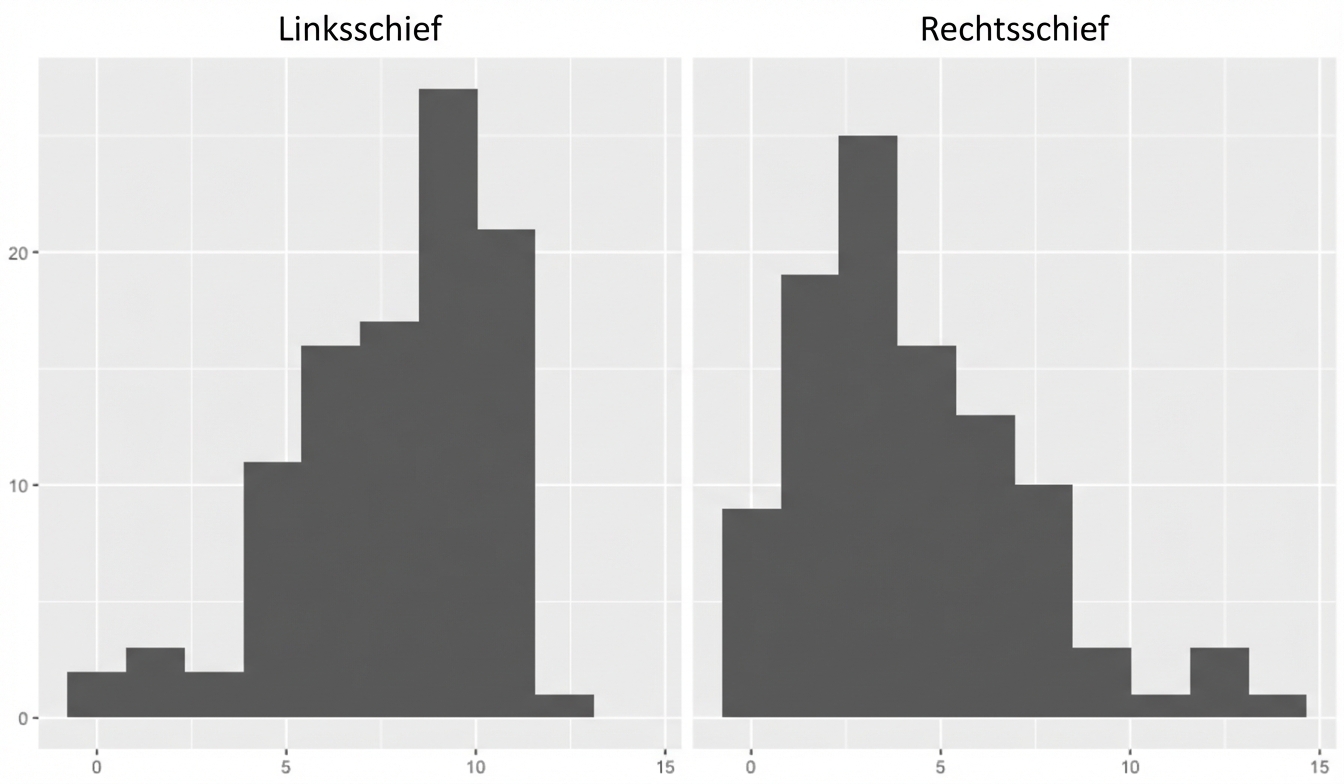
Welches Maß sollten wir verwenden?

