Properly Training a Model
Supervised Learning in R: Regression

Nina Zumel and John Mount
Win-Vector, LLC
Models can perform much better on training than they do on future data.
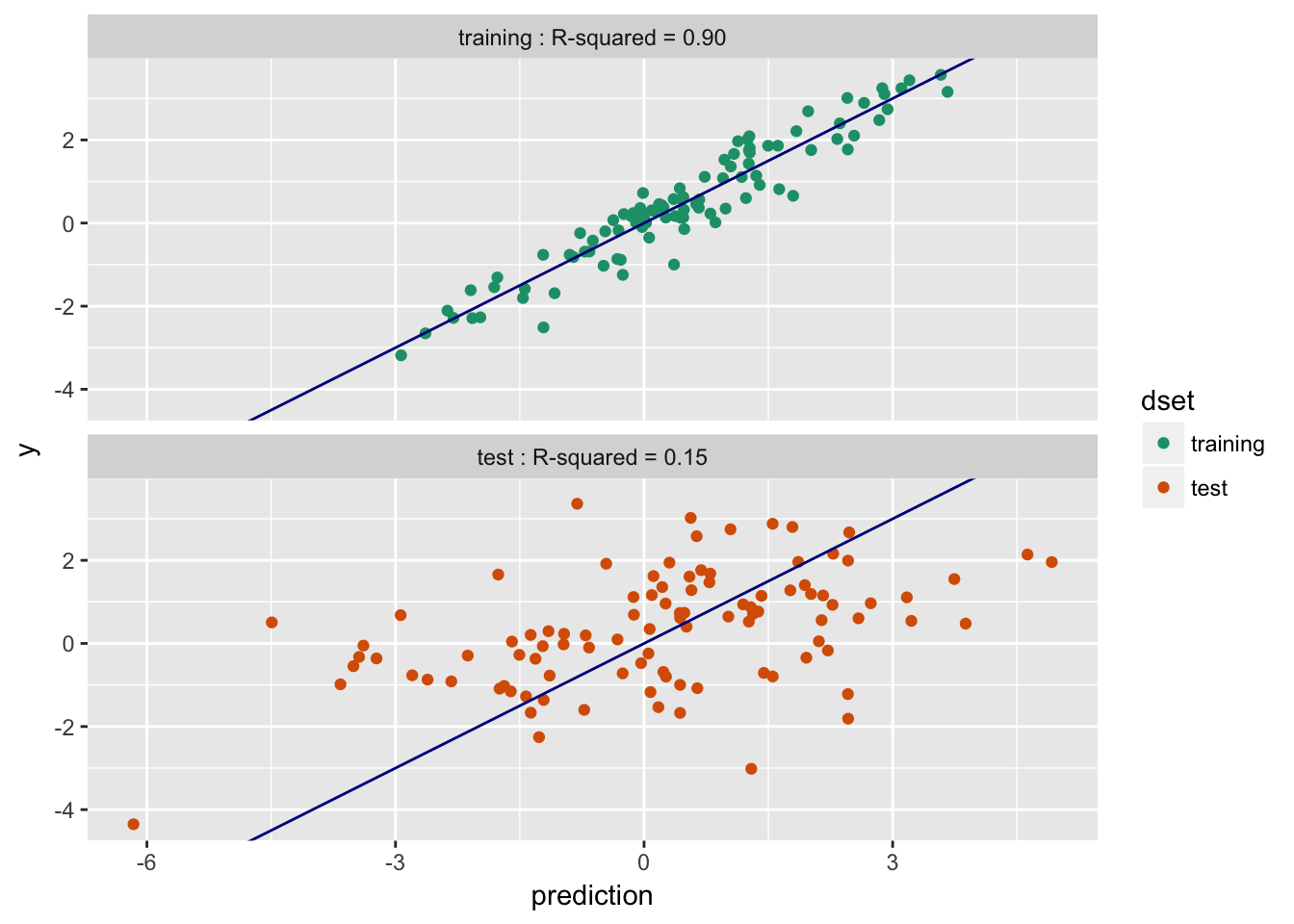
- Training $R^2$: 0.9; Test $R^2$: 0.15 -- Overfit
Test/Train Split
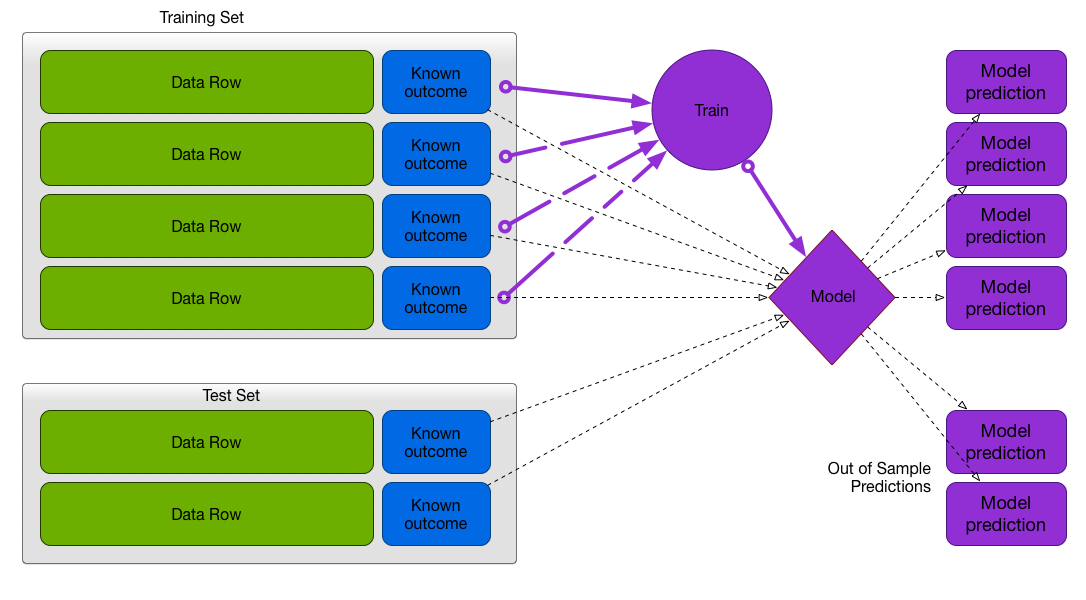
Recommended method when data is plentiful
Example: Model Female Unemployment
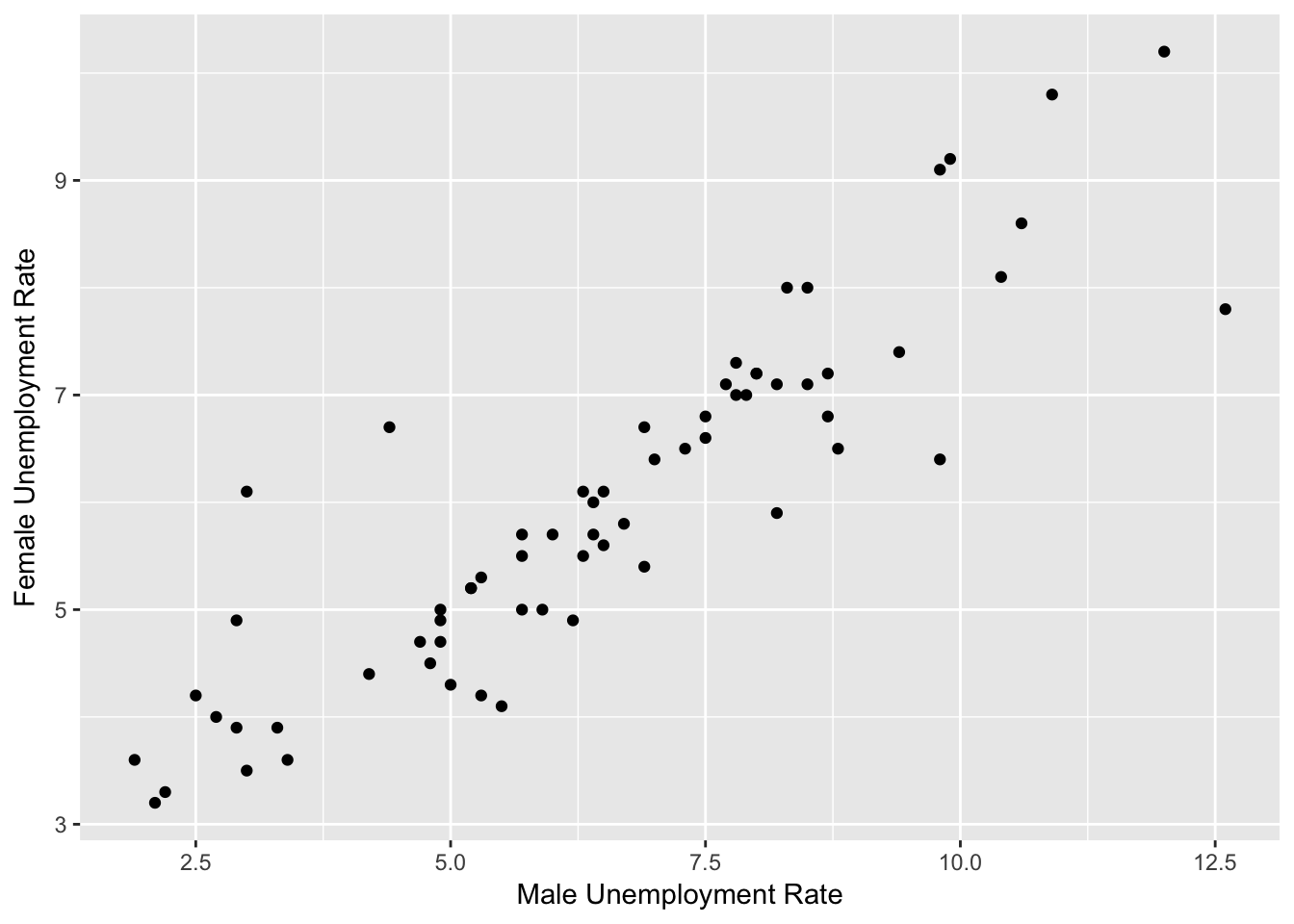
- Train on 66 rows, test on 30 rows
Model Performance: Train vs. Test
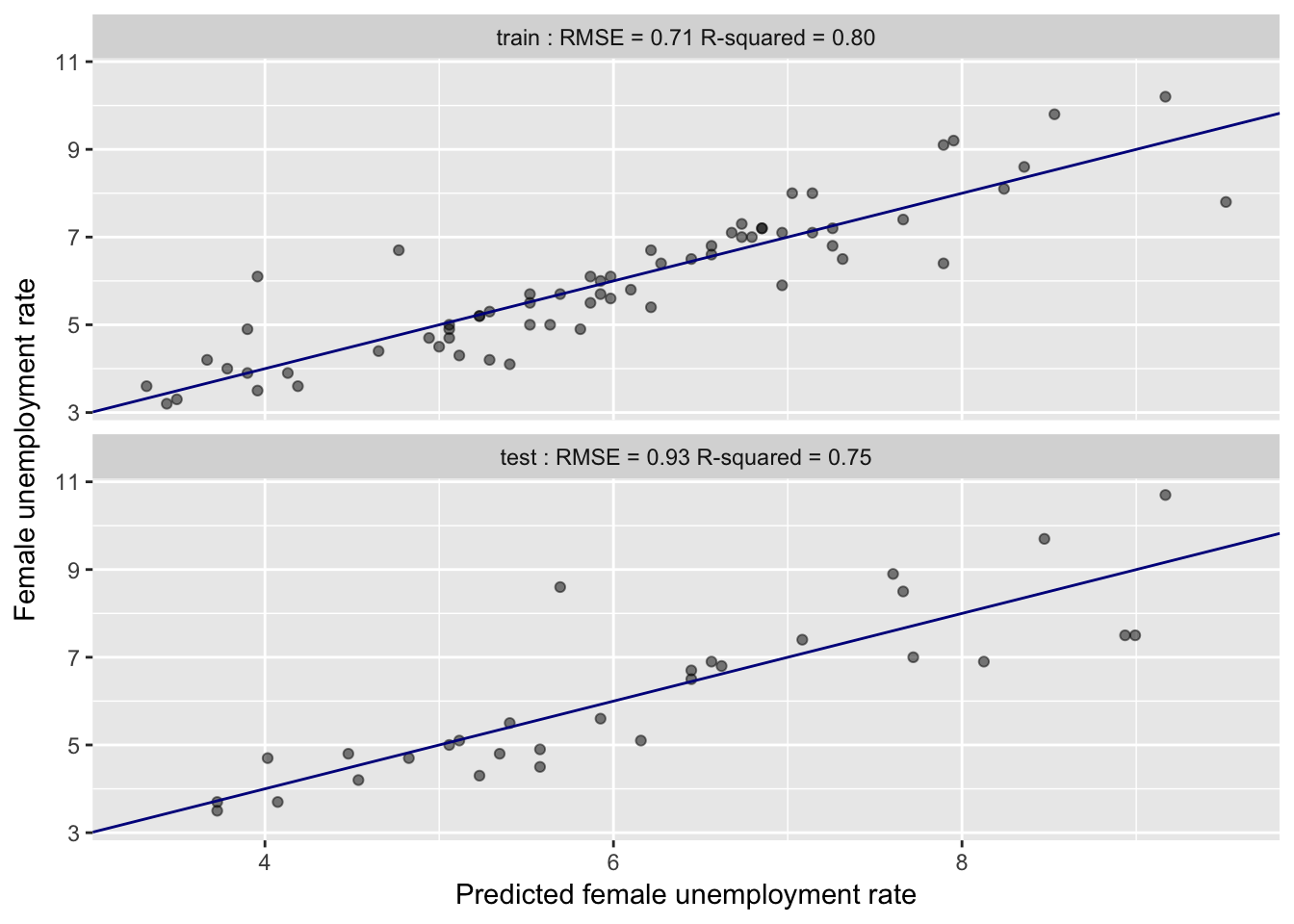
- Training: RMSE 0.71, $R^2$ 0.8
- Test: RMSE 0.93, $R^2$ 0.75
Cross-Validation
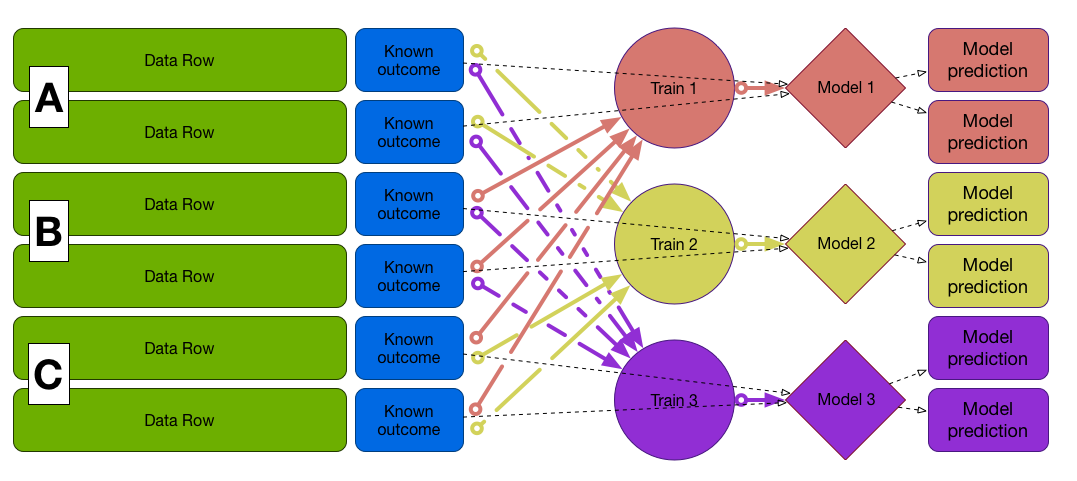
Preferred when data is not large enough to split off a test set
Cross-Validation
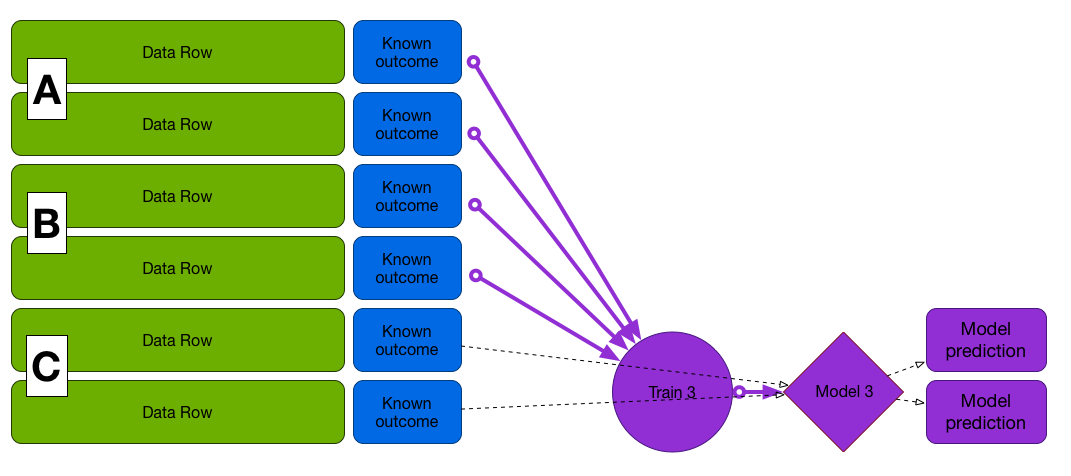
Cross-Validation
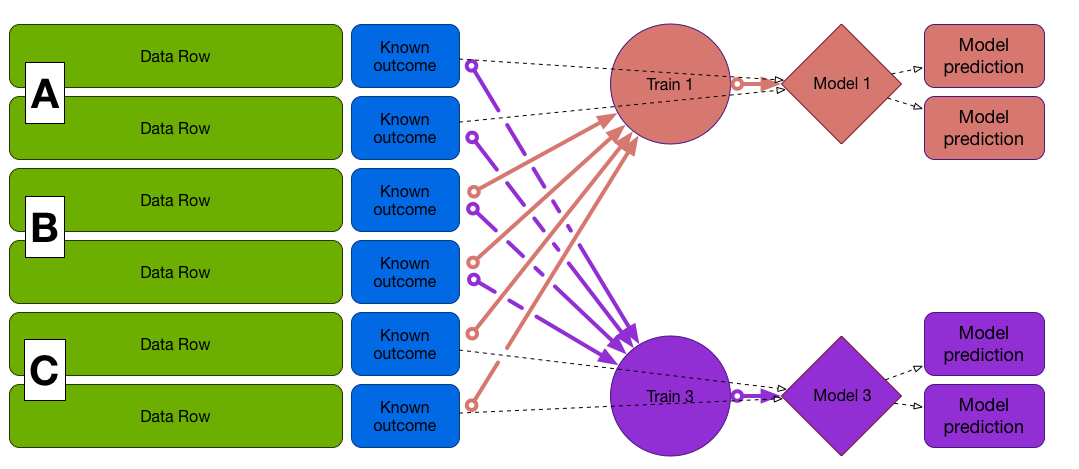
Cross-Validation

Create a cross-validation plan
library(vtreat)
splitPlan <- kWayCrossValidation(nRows, nSplits, NULL, NULL)
nRows: number of rows in the training datanSplits: number folds (partitions) in the cross-validation- e.g, nfolds = 3 for 3-way cross-validation
- remaining 2 arguments not needed here
Create a cross-validation plan
library(vtreat)
splitPlan <- kWayCrossValidation(10, 3, NULL, NULL)
First fold (A and B to train, C to test)
splitPlan[[1]]
$train
1 2 4 5 7 9 10
$app
3 6 8
Train on A and B, test on C, etc...
split <- splitPlan[[1]]
model <- lm(fmla, data = df[split$train,])
df$pred.cv[split$app] <- predict(model, newdata = df[split$app,])
Final Model
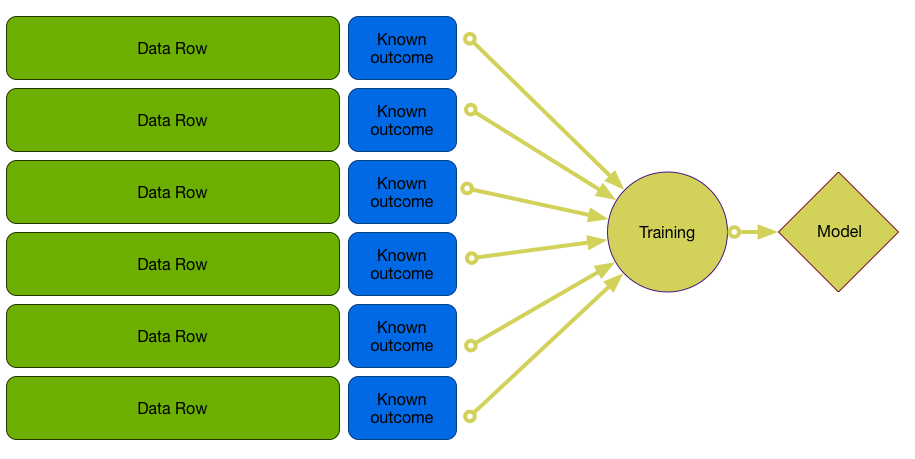
Example: Unemployment Model
| Measure type | RMSE | $R^2$ |
|---|---|---|
| train | 0.7082675 | 0.8029275 |
| test | 0.9349416 | 0.7451896 |
| cross-validation | 0.8175714 | 0.7635331 |
Let's practice!
Supervised Learning in R: Regression

