Logistic regression to predict probabilities
Supervised Learning in R: Regression

Nina Zumel and John Mount
Win-Vector LLC
Predicting Probabilities
- Predicting whether an event occurs (yes/no): classification
- Predicting the probability that an event occurs: regression
- Linear regression: predicts values in [$-\infty$, $\infty$]
- Probabilities: limited to [0,1] interval
- So we'll call it non-linear
Example: Predicting Duchenne Muscular Dystrophy (DMD)
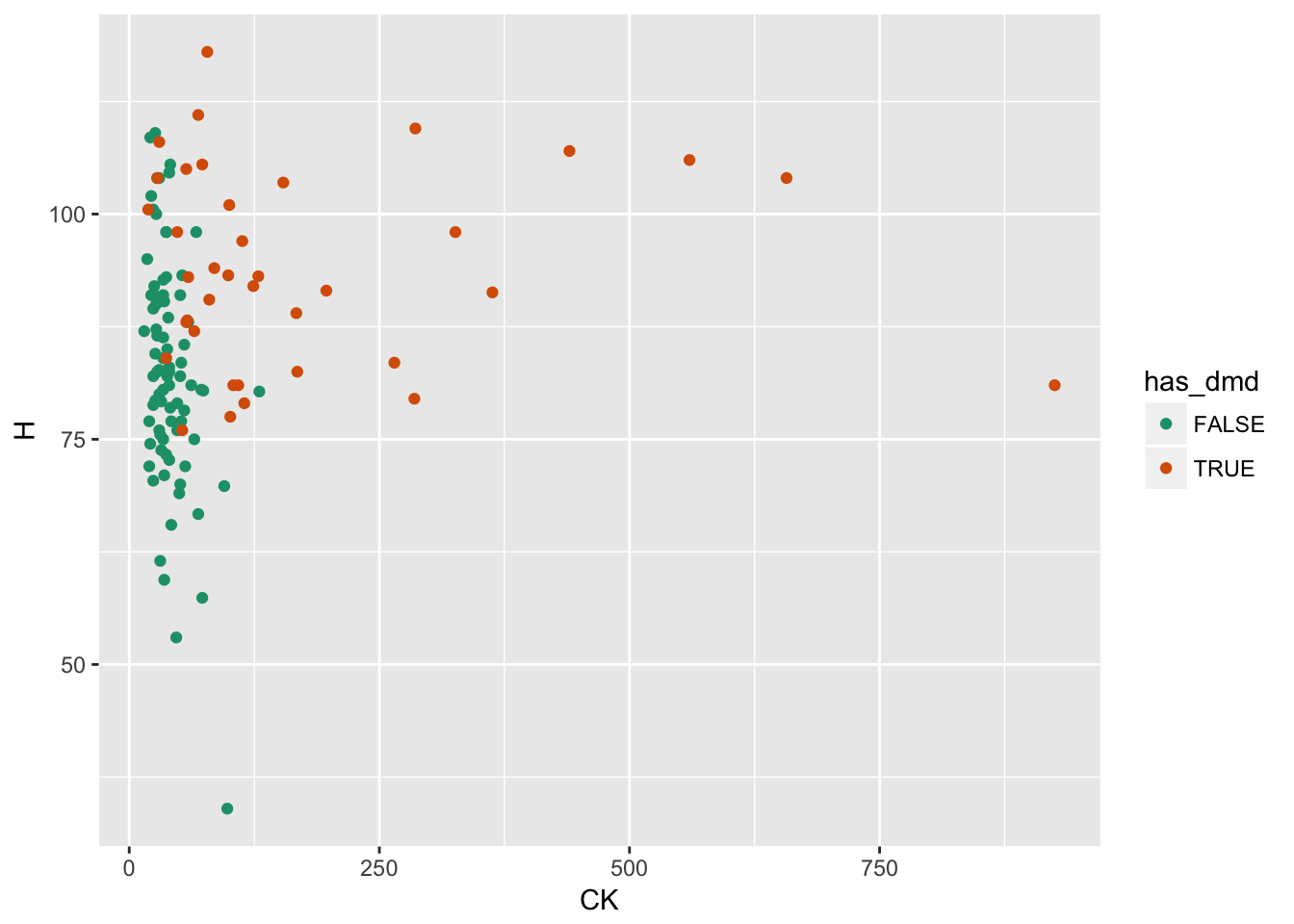
- outcome:
has_dmdinputs:CK,H
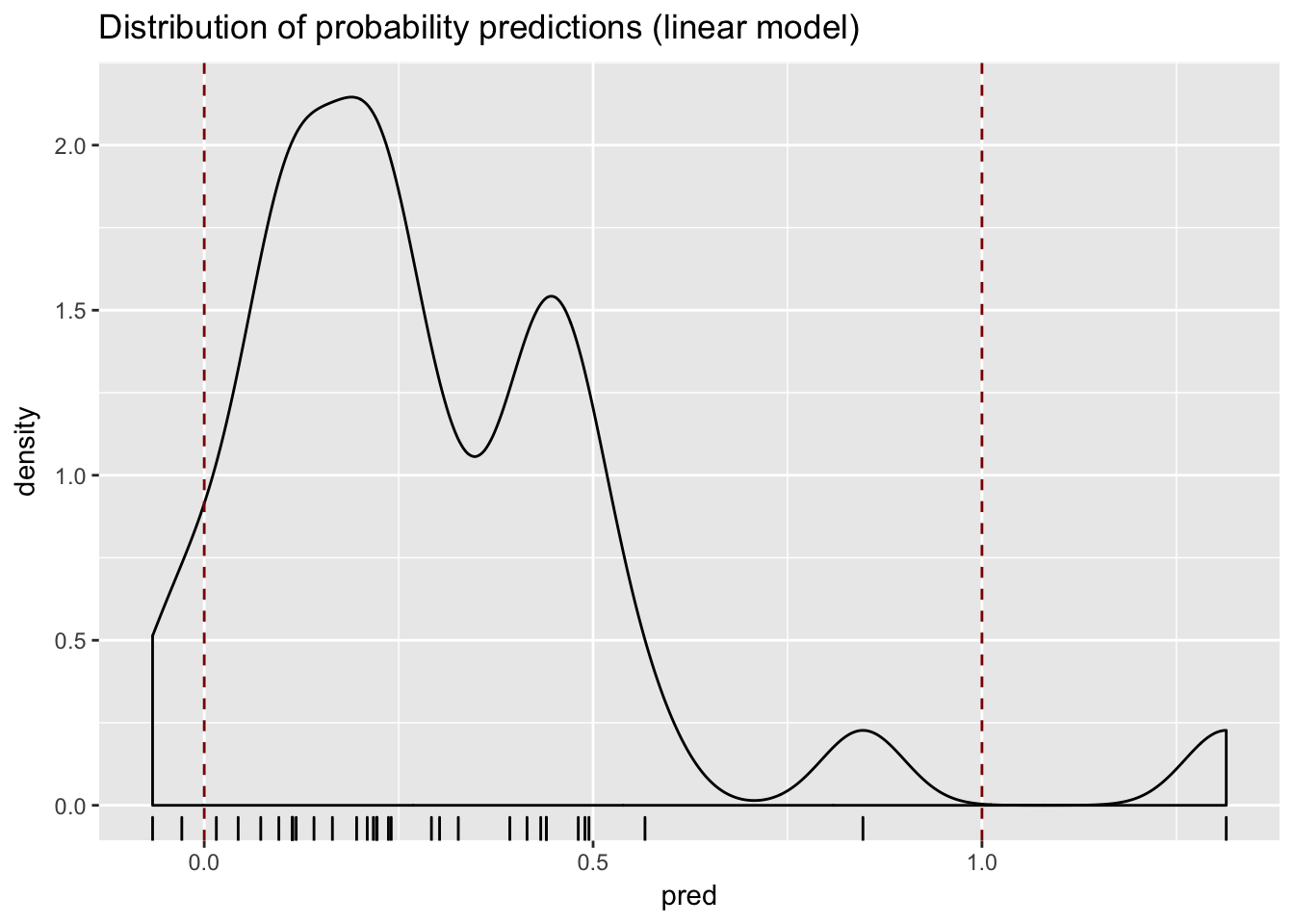
A Linear Regression Model
model <- lm(has_dmd ~ CK + H,
data = train)
test$pred <- predict(
model,
newdata = test
)
outcome: has_dmd $\in$ {0,1}
- 0: FALSE
- 1: TRUE
Model predicts values outside the range [0:1]
Logistic Regression
$$ log(\frac{p}{1-p}) = \beta_0 + \beta_1 x_1 + \beta_2 x_2 + ... $$
glm(formula, data, family = binomial)
- Generalized linear model
- Assumes inputs additive, linear in log-odds: $log( p/(1-p) )$
- family: describes error distribution of the model
- logistic regression:
family = binomial
- logistic regression:
DMD model
model <- glm(has_dmd ~ CK + H, data = train, family = binomial)
- outcome: two classes, e.g. $a$ and $b$
- model returns $Prob(b)$
- Recommend: 0/1 or FALSE/TRUE
Interpreting Logistic Regression Models
model
Call: glm(formula = has_dmd ~ CK + H, family = binomial, data = train)
Coefficients:
(Intercept) CK H
-16.22046 0.07128 0.12552
Degrees of Freedom: 86 Total (i.e. Null); 84 Residual
Null Deviance: 110.8
Residual Deviance: 45.16 AIC: 51.16
Predicting with a glm() model
predict(model, newdata, type = "response")
newdata: by default, training data- To get probabilities: use
type = "response"- By default: returns log-odds
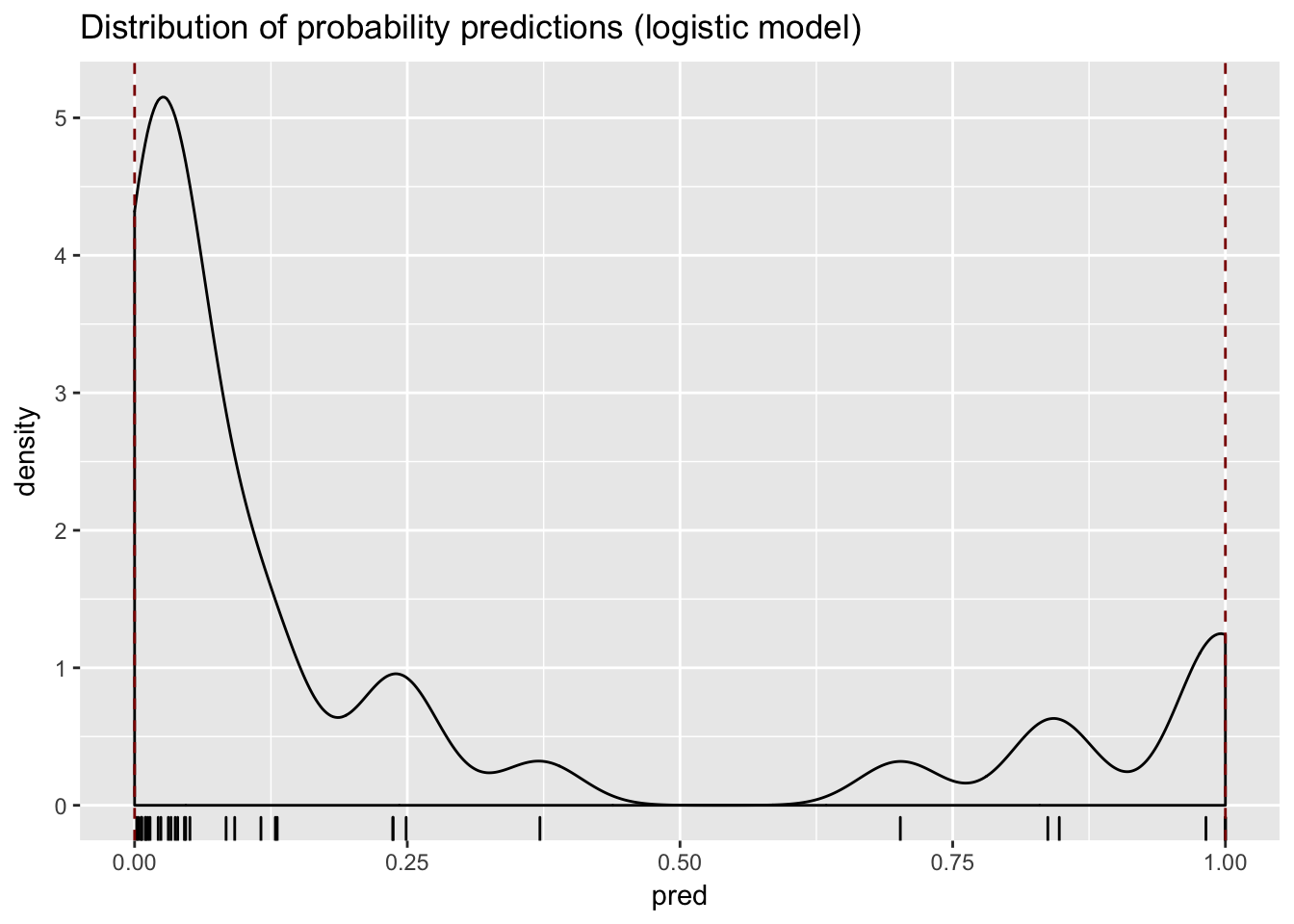
DMD Model
model <- glm(has_dmd ~ CK + H, data = train, family = binomial)
test$pred <- predict(model, newdata = test, type = "response")
Evaluating a logistic regression model: pseudo-$R^2$
$$ R^2 = 1 - \frac{RSS}{SS_{Tot}} $$
$$ pseudo R^2 = 1 - \frac{deviance}{null.deviance} $$
- Deviance: analogous to variance (RSS)
- Null deviance: Similar to $SS_{Tot}$
- pseudo R^2: Deviance explained
Pseudo-$R^2$ on Training data
Using broom::glance()
glance(model) %>%
summarize(pR2 = 1 - deviance/null.deviance)
pseudoR2
1 0.5922402
Using sigr::wrapChiSqTest()
wrapChiSqTest(model)
"... pseudo-R2=0.59 ..."
Pseudo-$R^2$ on Test data
# Test data
test %>%
mutate(pred = predict(model, newdata = test, type = "response")) %>%
wrapChiSqTest("pred", "has_dmd", TRUE)
Arguments:
- data frame
- prediction column name
- outcome column name
- target value (target event)
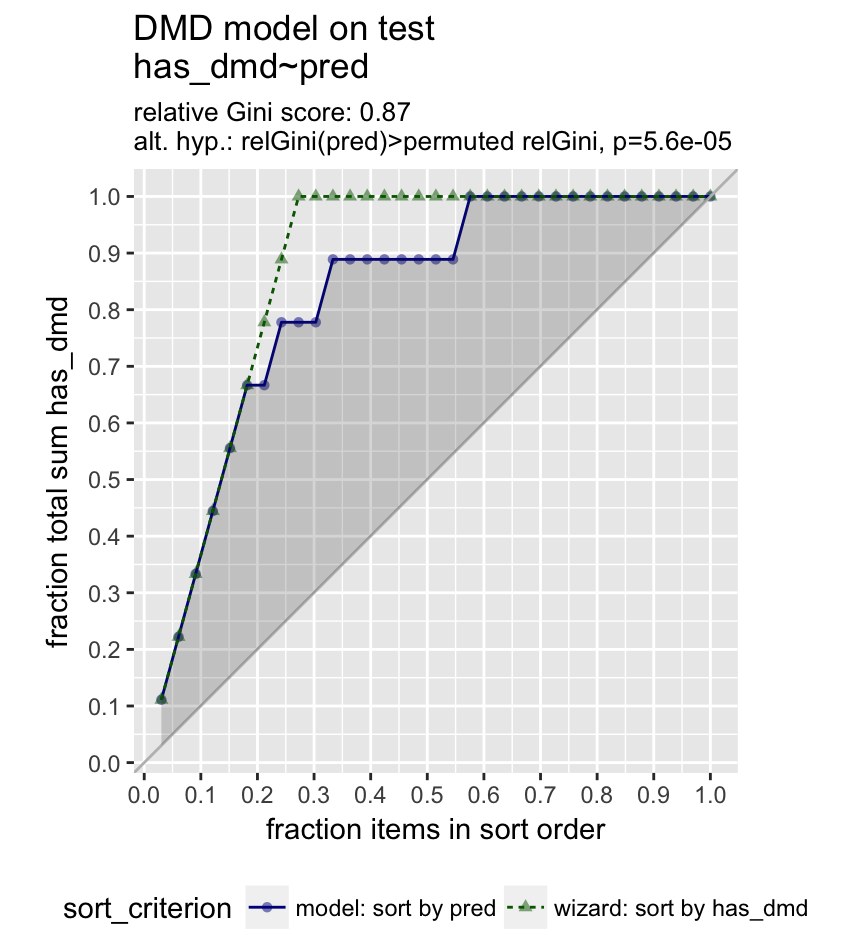
The Gain Curve Plot
GainCurvePlot(test, "pred","has_dmd", "DMD model on test")
Let's practice!
Supervised Learning in R: Regression

