Valeurs aberrantes, levier et influence
Introduction à la régression avec statsmodels en Python

Maarten Van den Broeck
Content Developer at DataCamp
Ensemble de données sur les gardons
roach = fish[fish['species'] == "Roach"]
print(roach.head())
species mass_g length_cm
35 Roach 40.0 12.9
36 Roach 69.0 16.5
37 Roach 78.0 17.5
38 Roach 87.0 18.2
39 Roach 120.0 18.6

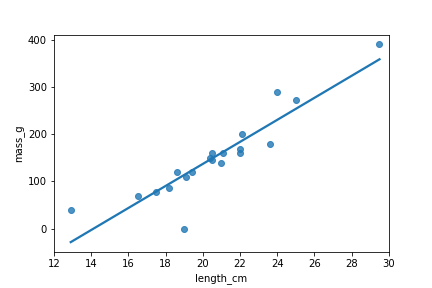
Quels points sont des valeurs aberrantes ?
sns.regplot(x="length_cm",
y="mass_g",
data=roach,
ci=None)
plt.show()
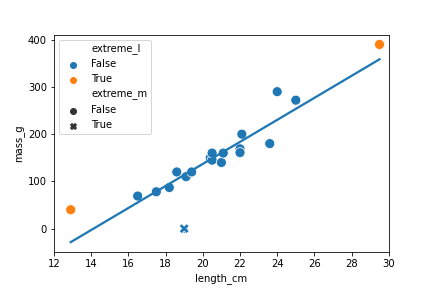
Valeurs explicatives extrêmes
roach["extreme_l"] = ((roach["length_cm"] < 15) |
(roach["length_cm"] > 26))
fig = plt.figure()
sns.regplot(x="length_cm",
y="mass_g",
data=roach,
ci=None)
sns.scatterplot(x="length_cm",
y="mass_g",
hue="extreme_l",
data=roach)
![Diagramme en nuage de points représentant la masse des gardons en fonction de leur longueur, avec une courbe de tendance. La plupart des points sont de couleur bleue, mais un gardon très court et un gardon très long ont des points de couleur orange. (https://assets.datacamp.com/production/repositories/5857/datasets/312d375df71d931d4787dcfb926b69e183bb3b3f/scatter-roach-mass-vs-length-extreme-length.png =110)
Valeurs de réponse éloignées de la ligne de régression
roach["extreme_m"] = roach["mass_g"] < 1
fig = plt.figure()
sns.regplot(x="length_cm",
y="mass_g",
data=roach,
ci=None)
sns.scatterplot(x="length_cm",
y="mass_g",
hue="extreme_l",
style="extreme_m",
data=roach)
Levier et influence
Le levier est une mesure de l'extrême variation des valeurs des variables explicatives.
L'influence mesure dans quelle mesure le modèle changerait si vous retiriez l'observation de l'ensemble de données lors de la modélisation.

.get_influence() et .summary_frame()
mdl_roach = ols("mass_g ~ length_cm", data=roach).fit()summary_roach = mdl_roach.get_influence().summary_frame()roach["leverage"] = summary_roach["hat_diag"] print(roach.head())
species mass_g length_cm leverage
35 Roach 40.0 12.9 0.313729
36 Roach 69.0 16.5 0.125538
37 Roach 78.0 17.5 0.093487
38 Roach 87.0 18.2 0.076283
39 Roach 120.0 18.6 0.068387
Distance de Cook
La distance de Cook est la mesure d'influence la plus couramment utilisée.
roach["cooks_dist"] = summary_roach["cooks_d"]
print(roach.head())
species mass_g length_cm leverage cooks_dist
35 Roach 40.0 12.9 0.313729 1.074015
36 Roach 69.0 16.5 0.125538 0.010429
37 Roach 78.0 17.5 0.093487 0.000020
38 Roach 87.0 18.2 0.076283 0.001980
39 Roach 120.0 18.6 0.068387 0.006610
Les gardons les plus influents
print(roach.sort_values("cooks_dist", ascending = False))
species mass_g length_cm leverage cooks_dist
35 Roach 40.0 12.9 0.313729 1.074015 # really short roach
54 Roach 390.0 29.5 0.394740 0.365782 # really long roach
40 Roach 0.0 19.0 0.061897 0.311852 # roach with zero mass
52 Roach 290.0 24.0 0.099488 0.150064
51 Roach 180.0 23.6 0.088391 0.061209
.. ... ... ... ... ...
43 Roach 150.0 20.4 0.050264 0.000257
44 Roach 145.0 20.5 0.050092 0.000256
42 Roach 120.0 19.4 0.056815 0.000199
47 Roach 160.0 21.1 0.050910 0.000137
37 Roach 78.0 17.5 0.093487 0.000020
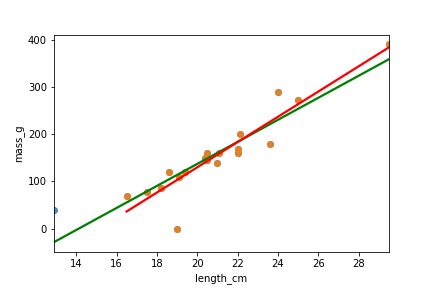
Suppression du gardon le plus influent
roach_not_short = roach[roach["length_cm"] != 12.9]
sns.regplot(x="length_cm",
y="mass_g",
data=roach,
ci=None,
line_kws={"color": "green"})
sns.regplot(x="length_cm",
y="mass_g",
data=roach_not_short,
ci=None,
line_kws={"color": "red"})
Passons à la pratique !
Introduction à la régression avec statsmodels en Python

