Introduction to differential binding
ChIP-seq with Bioconductor in R

Peter Humburg
Statistician, Macquarie University
Comparing samples
- Two groups of samples: Primary and treatment resistant tumor.
Questions we would like to answer:
- Are samples from the same group generally similar?
- Are samples from different groups different?
- What are those differences?
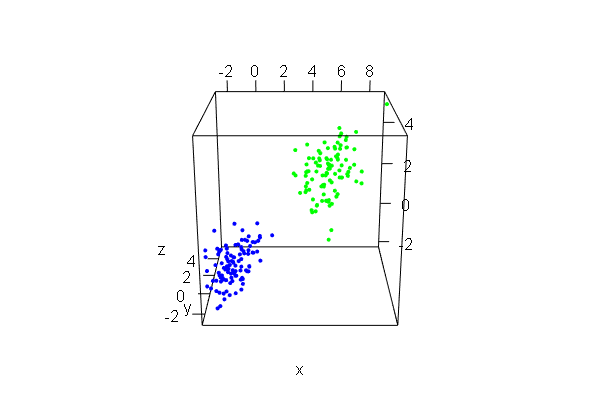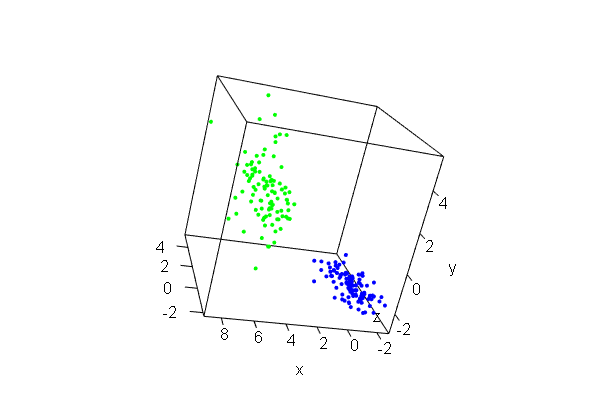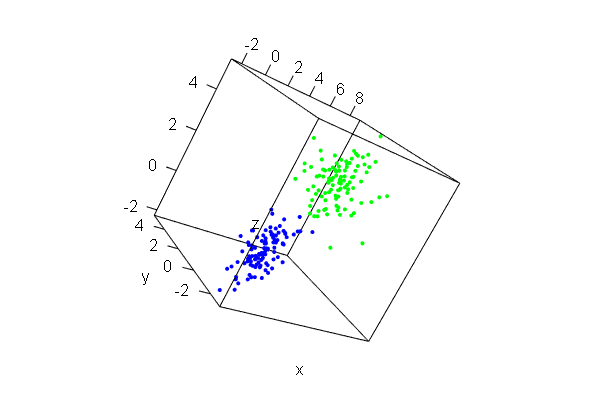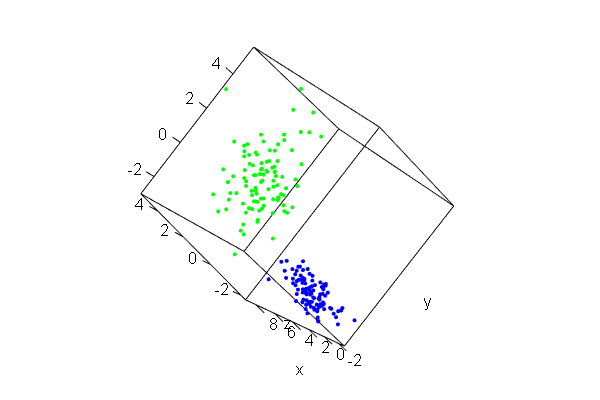
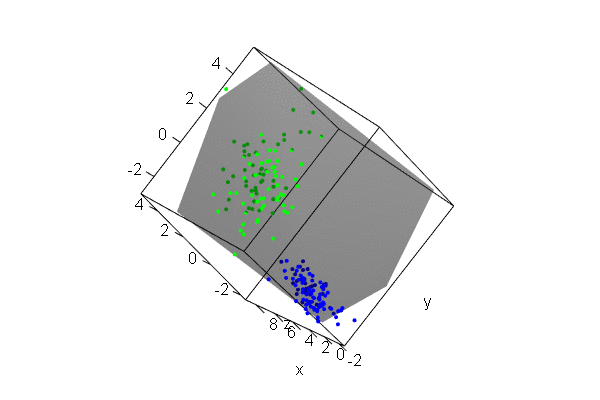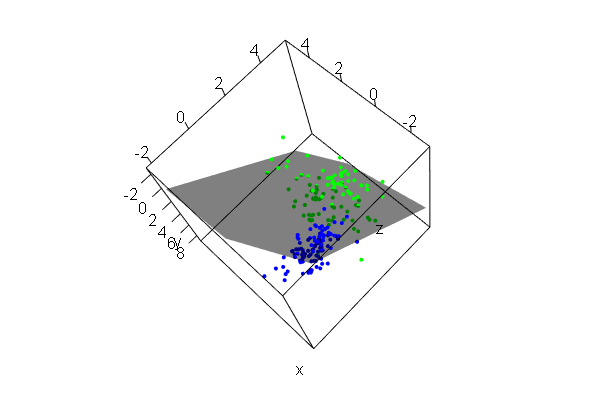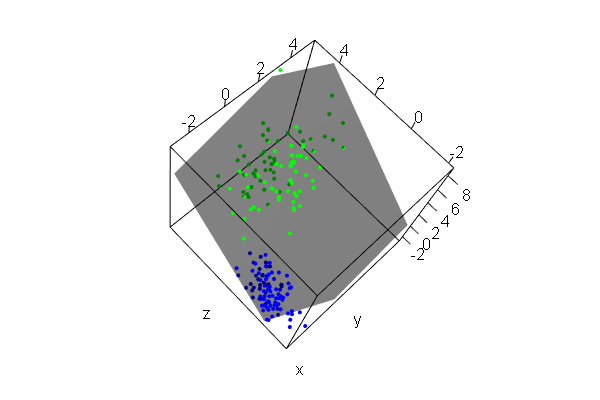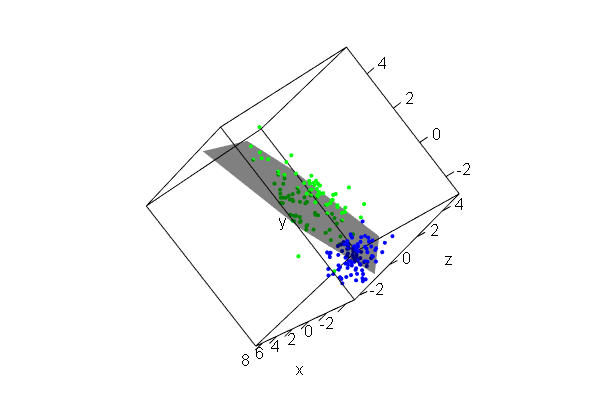
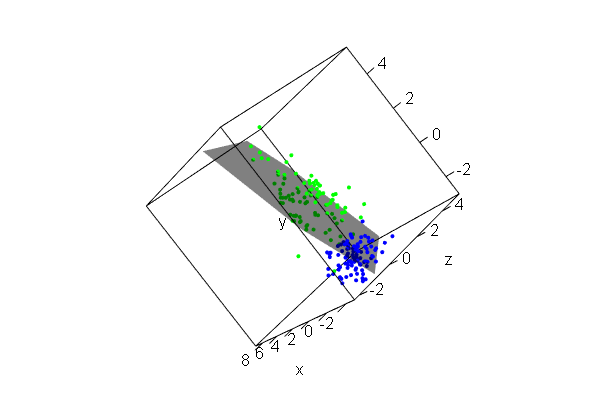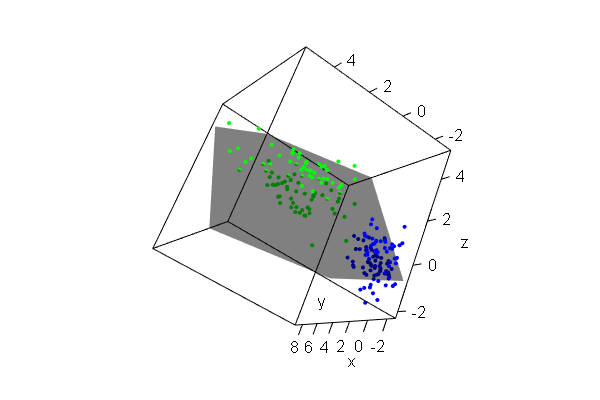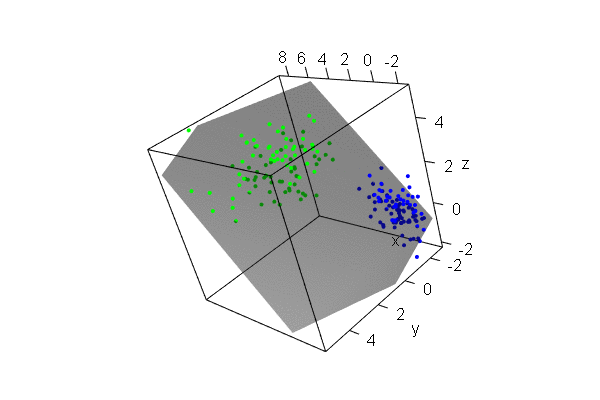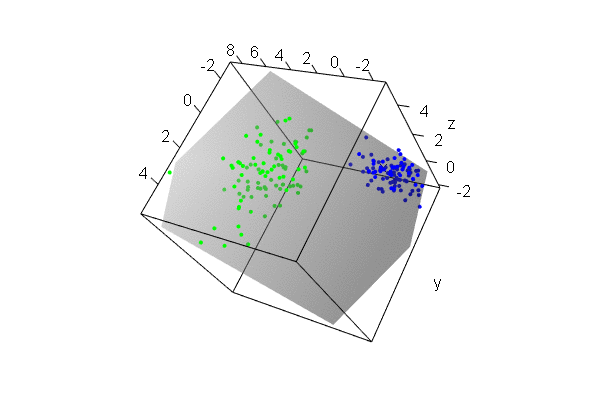
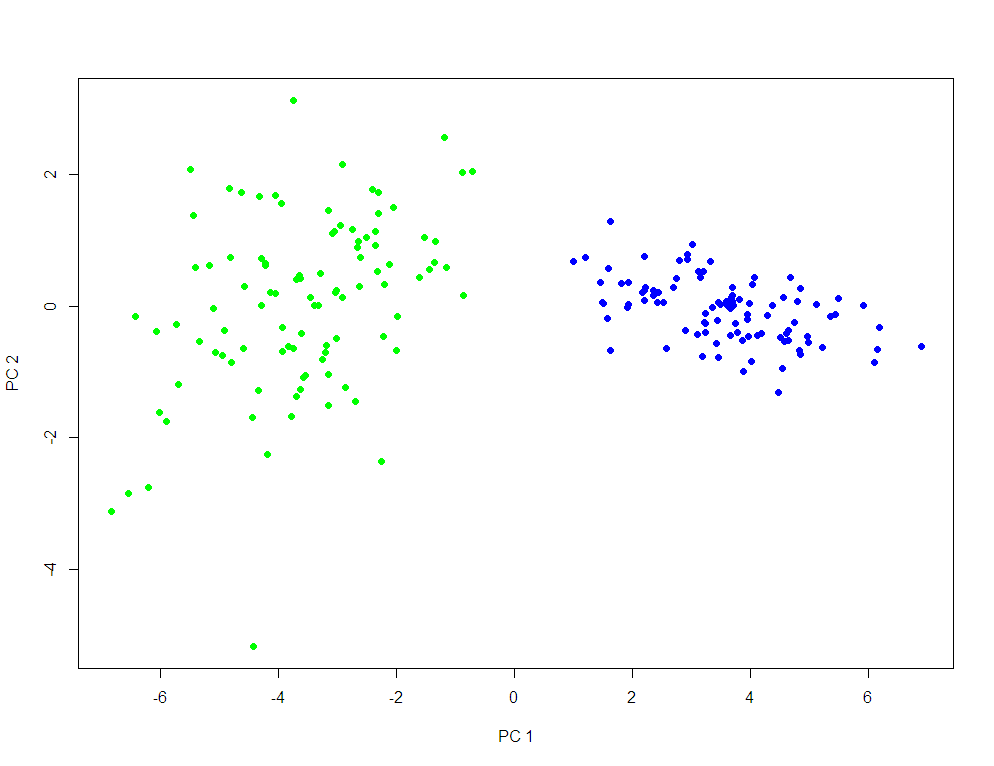
PCA plots for ChIP-seq data
qc_result <- ChIPQC("sample_info.csv", "hg19")
counts <- dba.count(qc_results, summits=250)
plotPrincomp(counts)
Hierarchical clustering
Compute distance between samples
distance <- dist(t(coverage))
Create dendogram
dendro <- hclust(distance)
Plot dendrogram
plot(dendro)
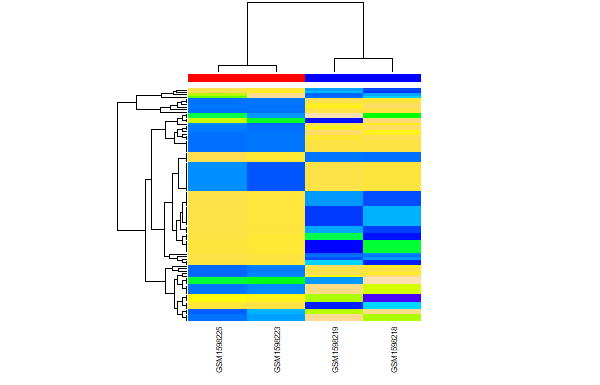
Heatmaps
Create heatmap from coverage data for peaks
dba.plotHeatmap(peaks, maxSites = peak_count, correlations = FALSE)
Let's practice!
ChIP-seq with Bioconductor in R

