Assessing enrichment
ChIP-seq with Bioconductor in R

Peter Humburg
Statistician, Macquarie University
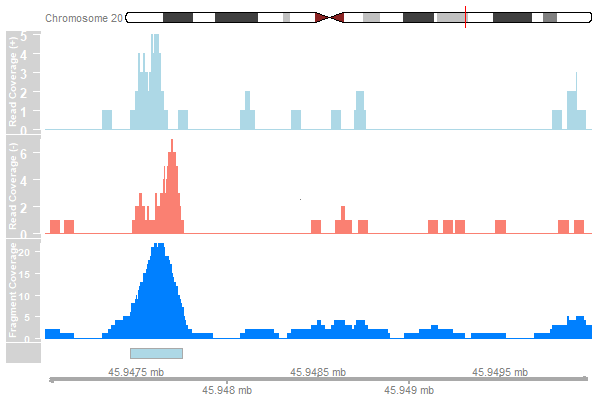
Extending reads
Load the data:
reads <- readGAlignments(bam)
reads_gr <- granges(reads[[1]])
Obtain average fragment length:
frag_length <- fragmentlength(qc_report)["GSM1598218"]
Extend reads and compute coverage:
reads_ext <- resize(reads_gr, width=frag_length)
cover_ext <- coverage(reads_ext)
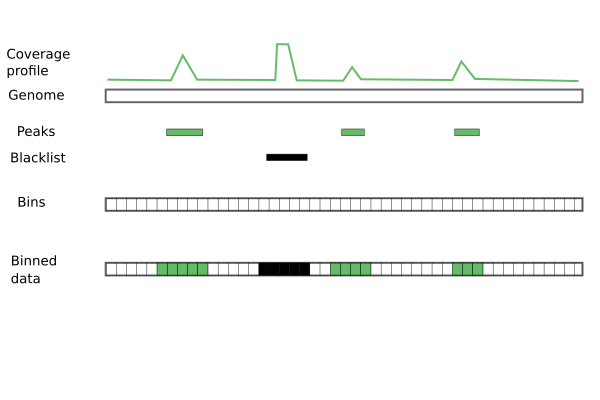
Coverage for peaks
Create 200 bp bins along the genome.
bins <- tileGenome(seqinfo(reads), tilewidth=200,
cut.last.tile.in.chrom=TRUE)
Find all bins overlapping peaks.
peak_bins_overlap <- findOverlaps(bins, peaks)
peak_bins <- bins[from(peak_bins_overlap), ]
Count the number of reads overlapping each peak bin.
peak_bins$score <- countOverlaps(peak_bins, reads)
Binned coverage function
count_bins <- function(reads, target, bins){
# Find all bins overlapping peaks
overlap <- from(findOverlaps(bins, target))
target_bins <- bins[overlap, ]
# Count the number of reads overlapping each peak bin
target_bins$score <- countOverlaps(target_bins, reads)
target_bins
}
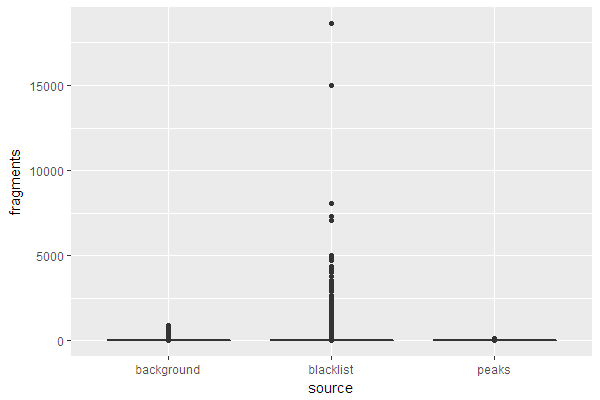
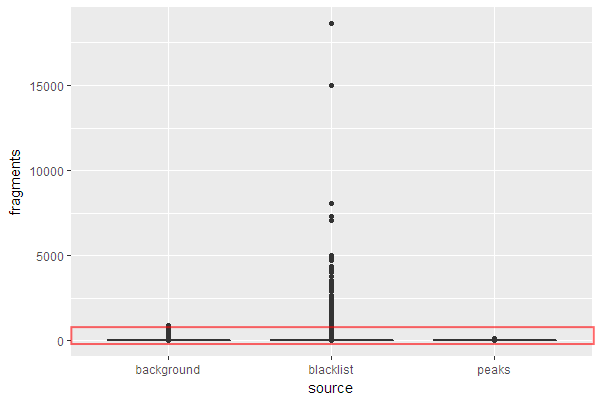
Coverage for blacklisted regions
peak_bins <- count_bins(reads_ext, peaks, bins)
bl_bins <- count_bins(reads_ext, blacklist.hg19, bins)
Background coverage
Remove all bins already accounted for.
bkg_bins <- subset(bins, !bins %in% peak_bins & !bins %in% bl_bins)
Count number of reads overlapping with each remaining bin.
bkg_bins$score <- countOverlaps(bkg_bins, reads_ext)
Let's practice!
ChIP-seq with Bioconductor in R

