Interpreting ChIP-seq peaks
ChIP-seq with Bioconductor in R

Peter Humburg
Statistician, Macquarie University
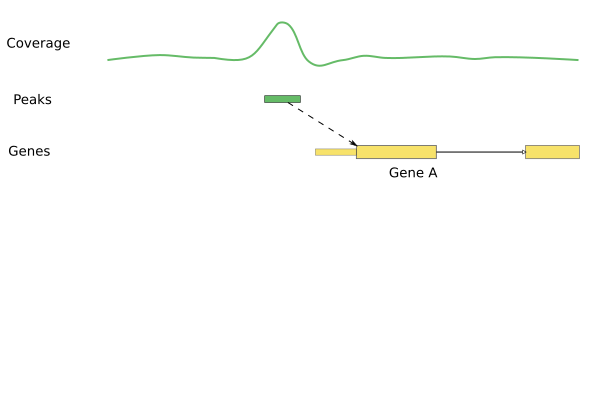
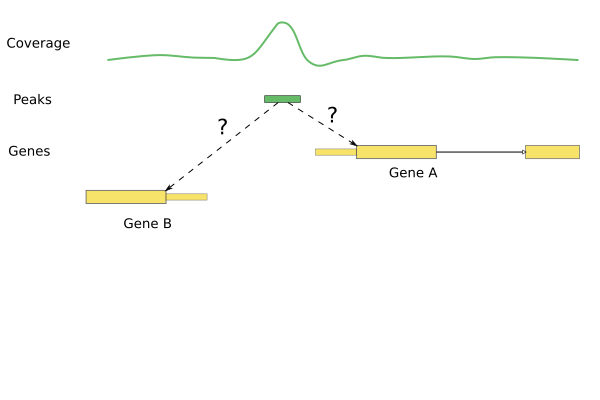
Annotating peaks
- Obtain information about gene locations.
- Assign peaks to genes.
- Identify genes associated with stronger peaks in one of the conditions.
library(TxDb.Hsapiens.UCSC.hg19.knownGene)
genes(TxDb.Hsapiens.UCSC.hg19.knownGene)
GRanges object with 23056 ranges and 1 metadata column:
seqnames ranges strand | gene_id
<Rle> <IRanges> <Rle> | <character>
1 chr19 [ 58858172, 58874214] - | 1
10 chr8 [ 18248755, 18258723] + | 10
100 chr20 [ 43248163, 43280376] - | 100
1000 chr18 [ 25530930, 25757445] - | 1000
10000 chr1 [243651535, 244006886] - | 10000
... ... ... ... . ...
9991 chr9 [114979995, 115095944] - | 9991
9992 chr21 [ 35736323, 35743440] + | 9992
9993 chr22 [ 19023795, 19109967] - | 9993
9994 chr6 [ 90539619, 90584155] + | 9994
9997 chr22 [ 50961997, 50964905] - | 9997
-------
seqinfo: 93 sequences (1 circular) from hg19 genome
Additional Annotations
library(org.Hs.eg.db)
select(org.Hs.eg.db, keys=gene_id,
columns="SYMBOL", keytype="ENTREZID")
ENTREZID SYMBOL
1 1 A1BG
2 10 NAT2
3 100 ADA
4 1000 CDH2
5 10000 AKT3
6 100008586 GAGE12F
...
Annotating Peaks
library(ChIPpeakAnno)annoPeaks(peaks, human_genes, bindingType="startSite", bindingRegion=c(-5000,5000))
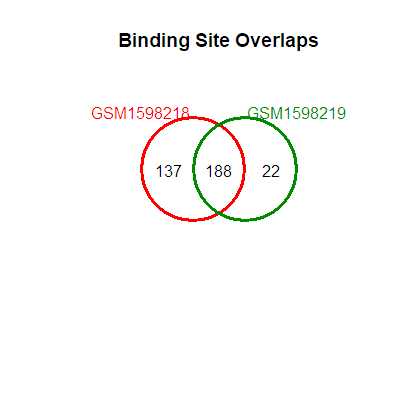
Visualizing similarities and differences
library(DiffBind)
dba.plotVenn(peaks, mask=1:2)
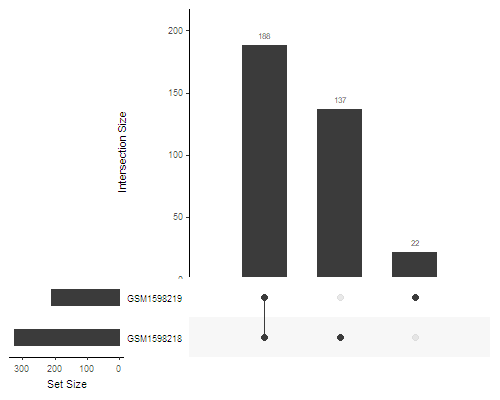
Using UpSet plots
library(UpSetR)
called_peaks <- as.data.frame(peaks$called)
upset(called_peaks, sets=colnames(peaks$called), order.by='freq')
Let's practice!
ChIP-seq with Bioconductor in R

