What is ChIP-seq?
ChIP-seq with Bioconductor in R

Peter Humburg
Statistician, Macquarie University
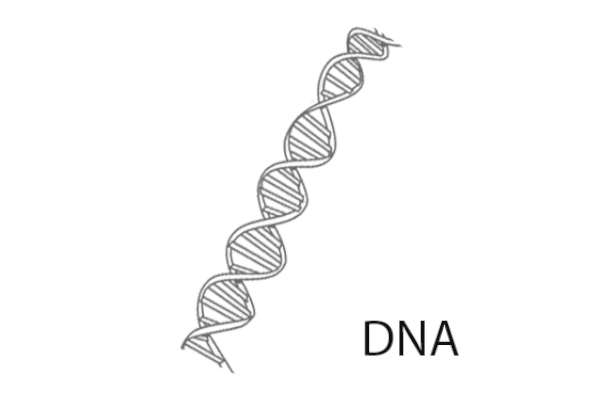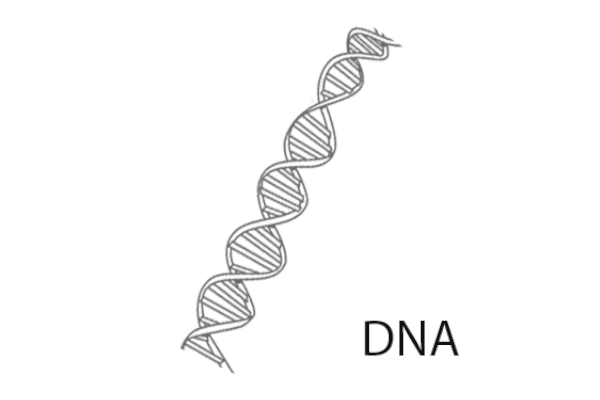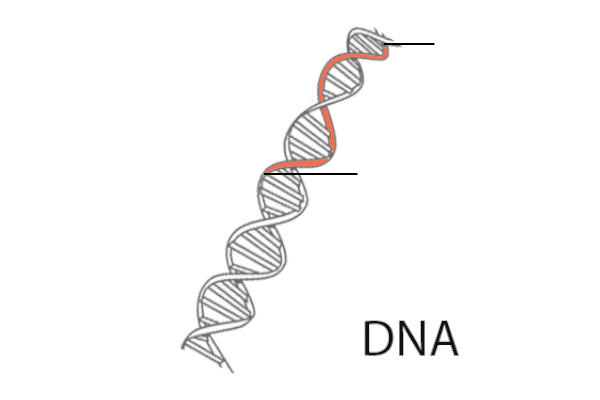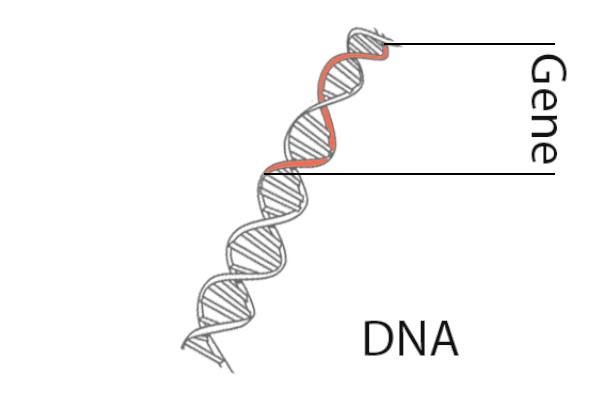

The Data
- Prostate cancer patients
- 5 primary tumor samples
- 3 treatment resistant samples
- ChIP targetting Androgen Receptor
- AR is activated by binding testosterone
- Activated AR regulates gene expression of target genes
Accessing ChIP-seq data in R
- Loading BAM files
library(GenomicAlignments)
reads <- readGAlignments('file_name')
- Obtaining read coordinates
seqnames(reads)
start(reads)
end(reads)
- Computing coverage
coverage(reads)
Accessing peak calls
- Loading BED files
library(rtracklayer)
peaks <- import.bed('file_name')
- Obtaining peak coordinates
chrom(peaks)
ranges(peaks)
- Extracting peak scores
score(peaks)
Let's practice!
ChIP-seq with Bioconductor in R

