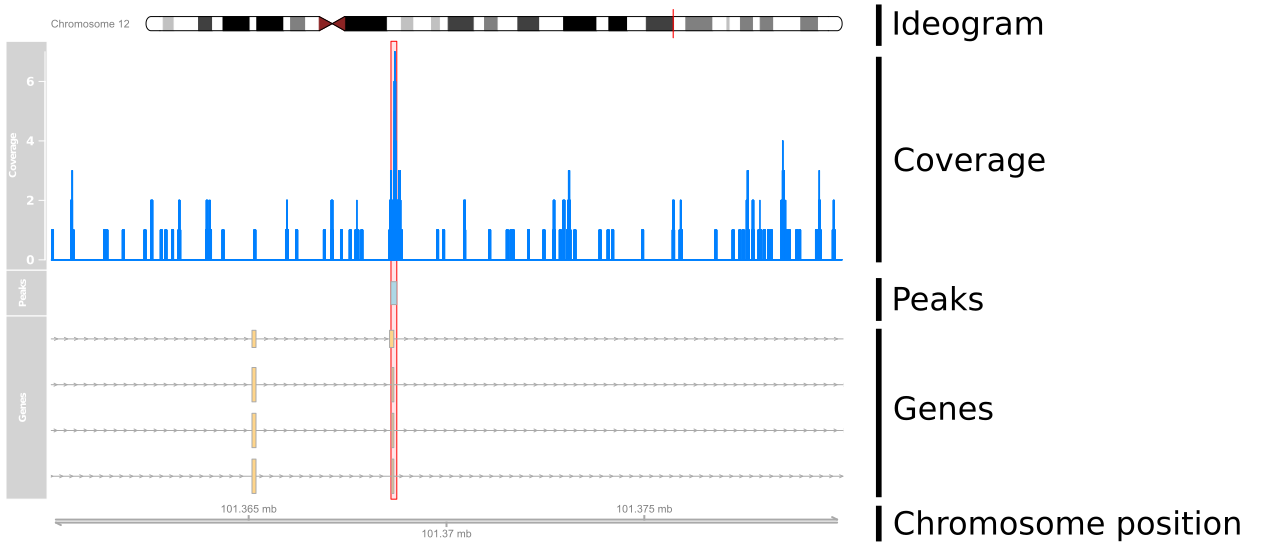
Taking a closer look at peaks
ChIP-seq with Bioconductor in R

Peter Humburg
Statistician, Macquarie University
Using Gviz
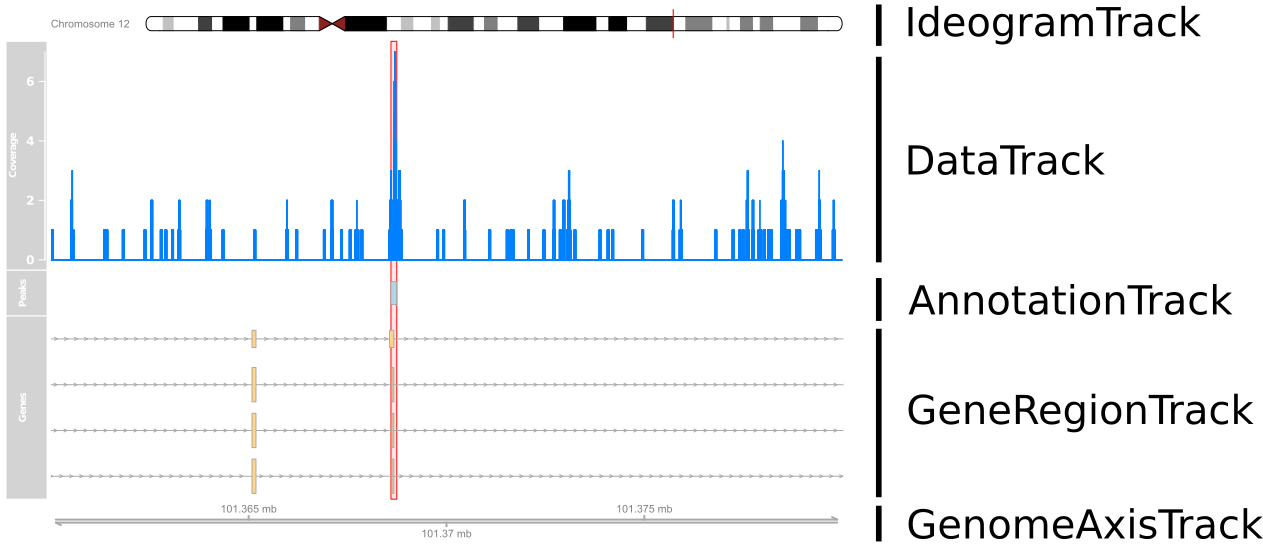
What are we plotting?
What are we plotting?
What are we plotting?
What are we plotting?
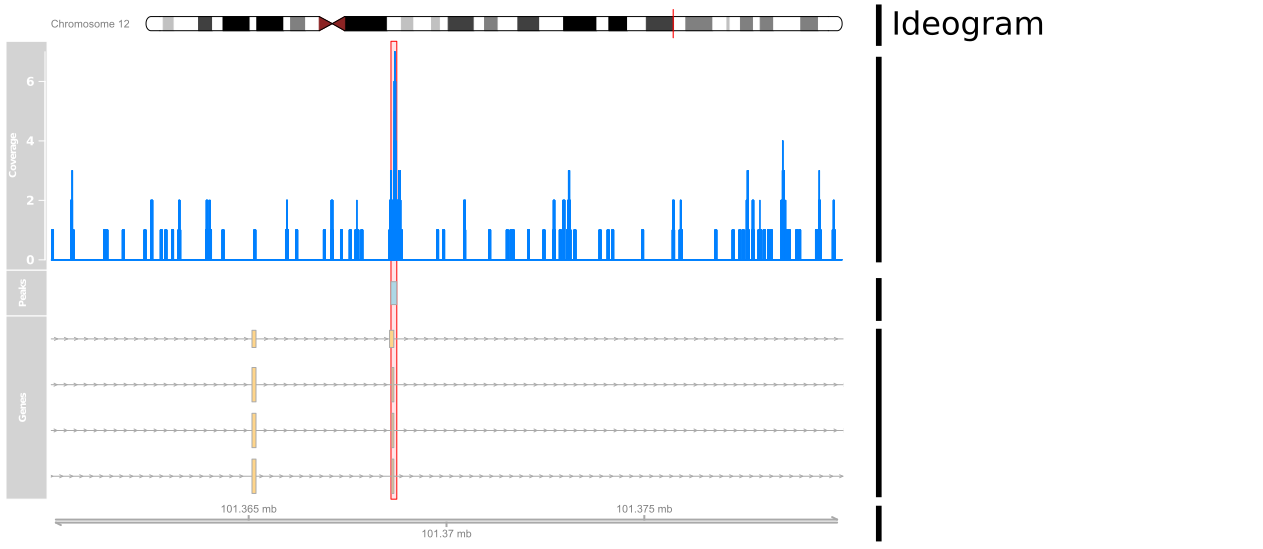
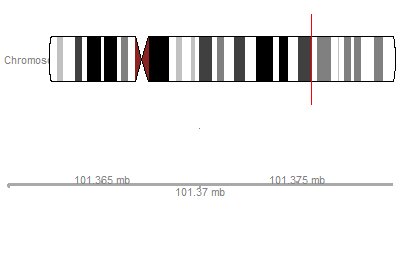
Setting-up coordinates
library(Gviz)ideogram <- IdeogramTrack("chr12", "hg19")axis <- GenomeAxisTrack()plotTracks(list(ideogram, axis), from=101360000, to=101380000)
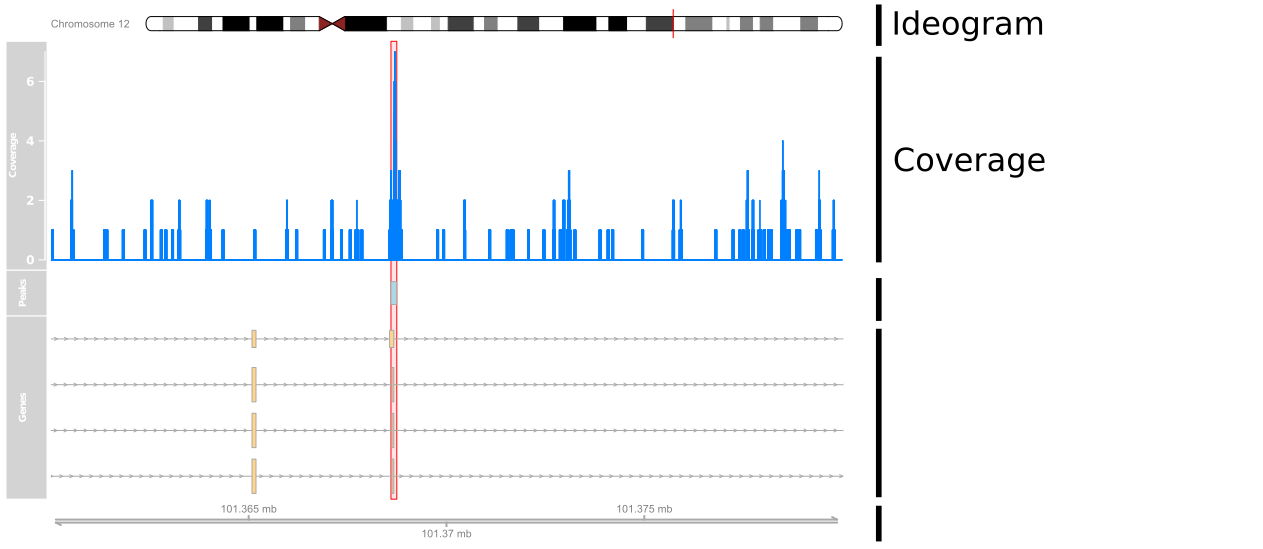
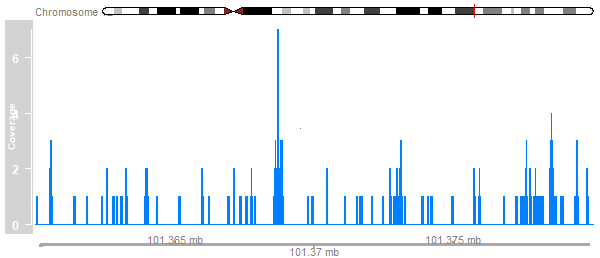
Adding Data
cover_track <- DataTrack(cover_ranges,window=100000,type='h',name="Coverage")
plotTracks(list(ideogram, cover_track, axis), from=101360000, to=101380000)
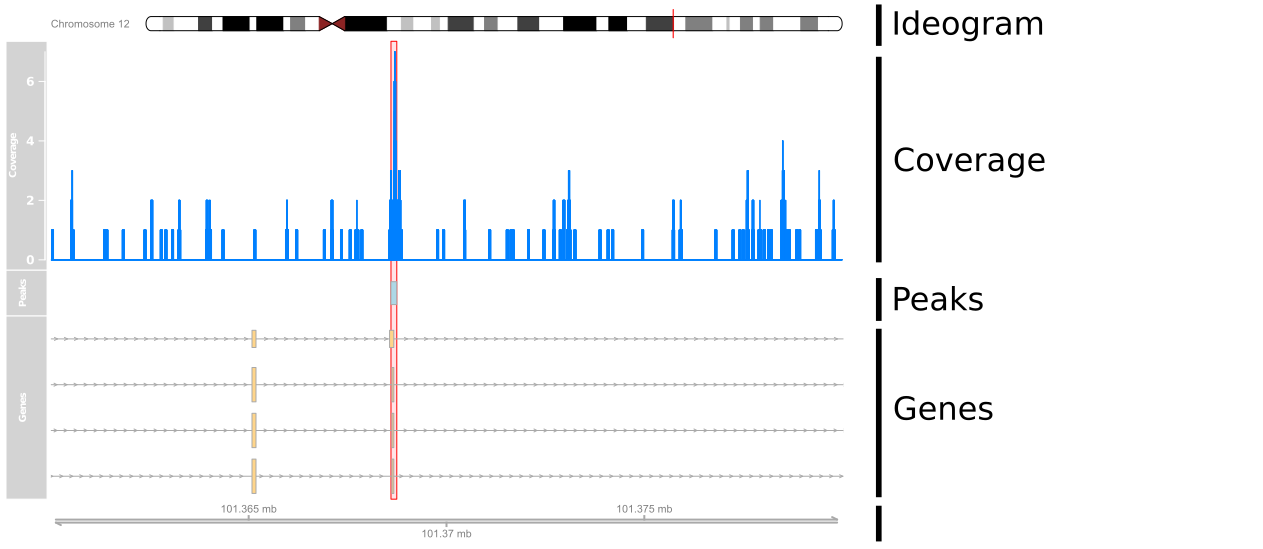
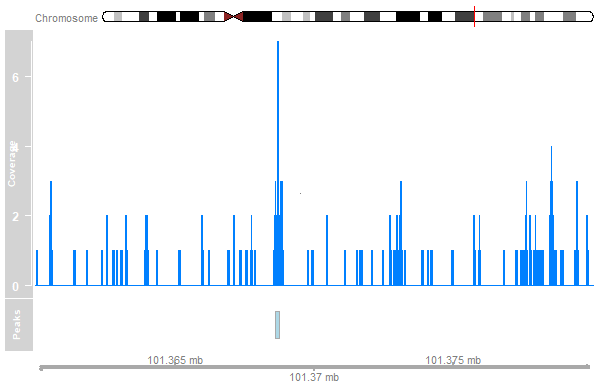
Adding Annotations
peak_track <- AnnotationTrack(peaks, name="Peaks")
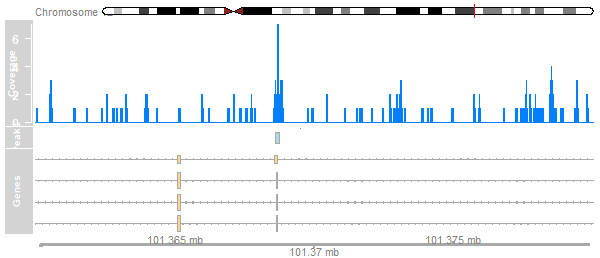
plotTracks(list(ideogram, cover_track, peak_track, axis),
from=101360000, to=101380000)
Gene Annotations
library(TxDb.Hsapiens.UCSC.hg19.knownGene)tx <- GeneRegionTrack(TxDb.Hsapiens.UCSC.hg19.knownGene, chromosome="chr12", start=101360000, end=101380000, name="Genes") plotTracks(list(ideogram, cover_track, peak_track, tx, axis), from=101360000, to=101380000)
Let's practice!
ChIP-seq with Bioconductor in R

