Importing data
ChIP-seq with Bioconductor in R

Peter Humburg
Statistician, Macquarie University
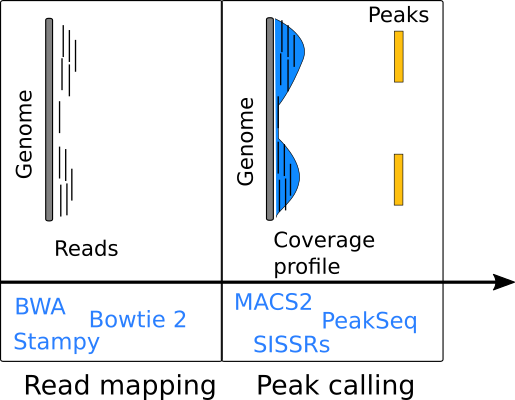
Handling sequence reads
- Usually stored in Binary Sequence Alignment/Map (BAM) format files.
- BAM record fields:
- Read name:
SRR1782620.7265769 - Binary flag:
0 - Reference sequence name and position of alignment:
chr20 29803915 - Mapping quality:
0 - CIGAR string (alignment summary):
51M - Reference sequence and position of paired read (not used here):
0 0 - Read sequence:
AATGAAATGGAA... - Read quality (ASCII encoded):
CCCFFFFFHHHH...
- Read name:
Importing mapped reads into R
- Use
Rsamtoolspackage to interact with BAM files. Rsamtoolsprovides functions for indexing, reading, filtering and writing of BAM files.
Use readGAlignments to import mapped reads.
library(GenomicAlignments)
reads <- readGAlignments(bam_file)
Returns GAlignments object.
Importing selected regions
- Use
BamViewsto define regions of interest.
library(GenomicRanges)
library(Rsamtools)
ranges <- GRanges(...)
views <- BamViews(bam_file, bamRanges=ranges)
- Then import reads as before.
reads <- readGAlignments(views)
The BamViews function supports multiple BAM files.
Importing peak calls
Use import.bed to load peak calls from a BED file.
library(rtracklayer)
peaks <- import.bed(peak_bed, genome="hg19")
Use peaks to define views into the BAM files.
bams <- BamViews(bam_file, bamRanges=peaks)
reads <- readGAlignments(bams)
Let's practice!
ChIP-seq with Bioconductor in R

