ChIP-seq Review
ChIP-seq with Bioconductor in R

Peter Humburg
Statistician, Macquarie University
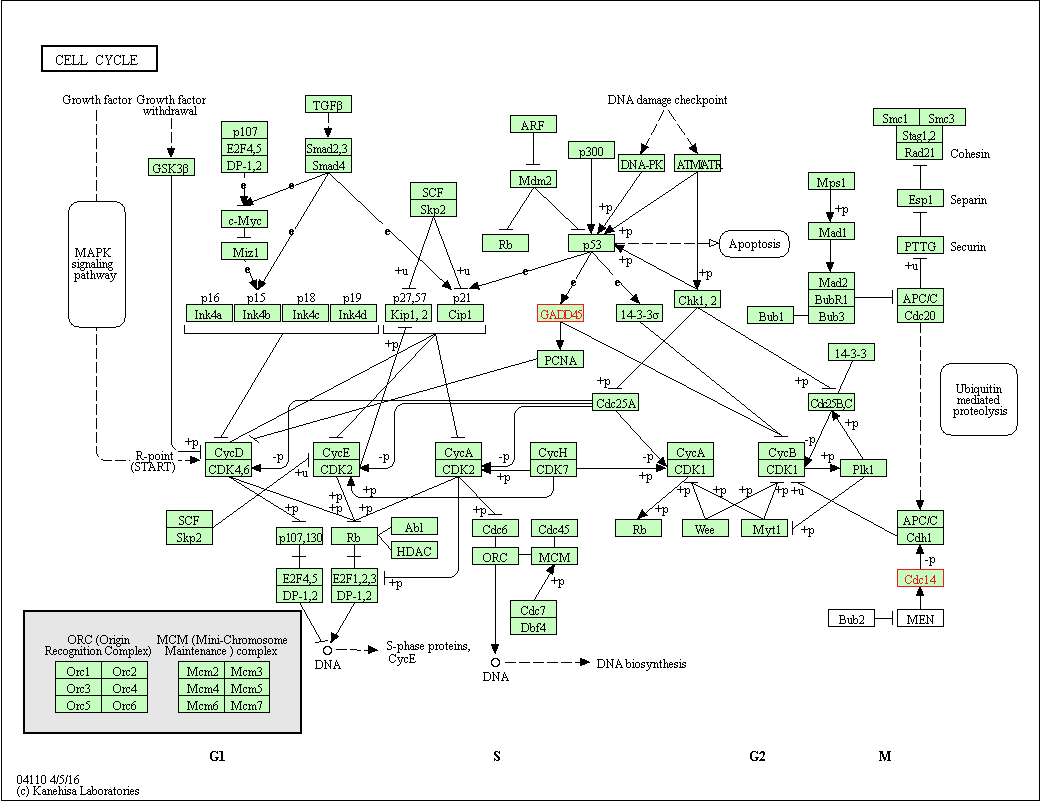
Review: Importing data
Load peak calls
peaks <- import.bed(peak_file, genome="hg19")
Create a BamViews object
bam_views <- BamViews(bam_file, bamRanges=peaks)
Load the reads
reads <- readGAlignments(bam_views)
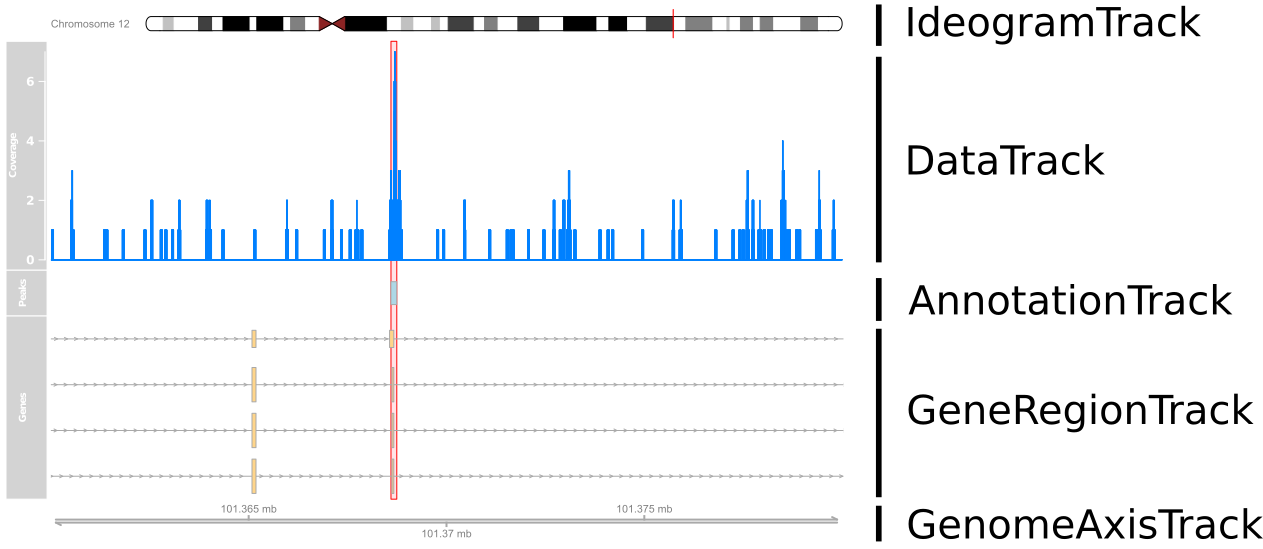
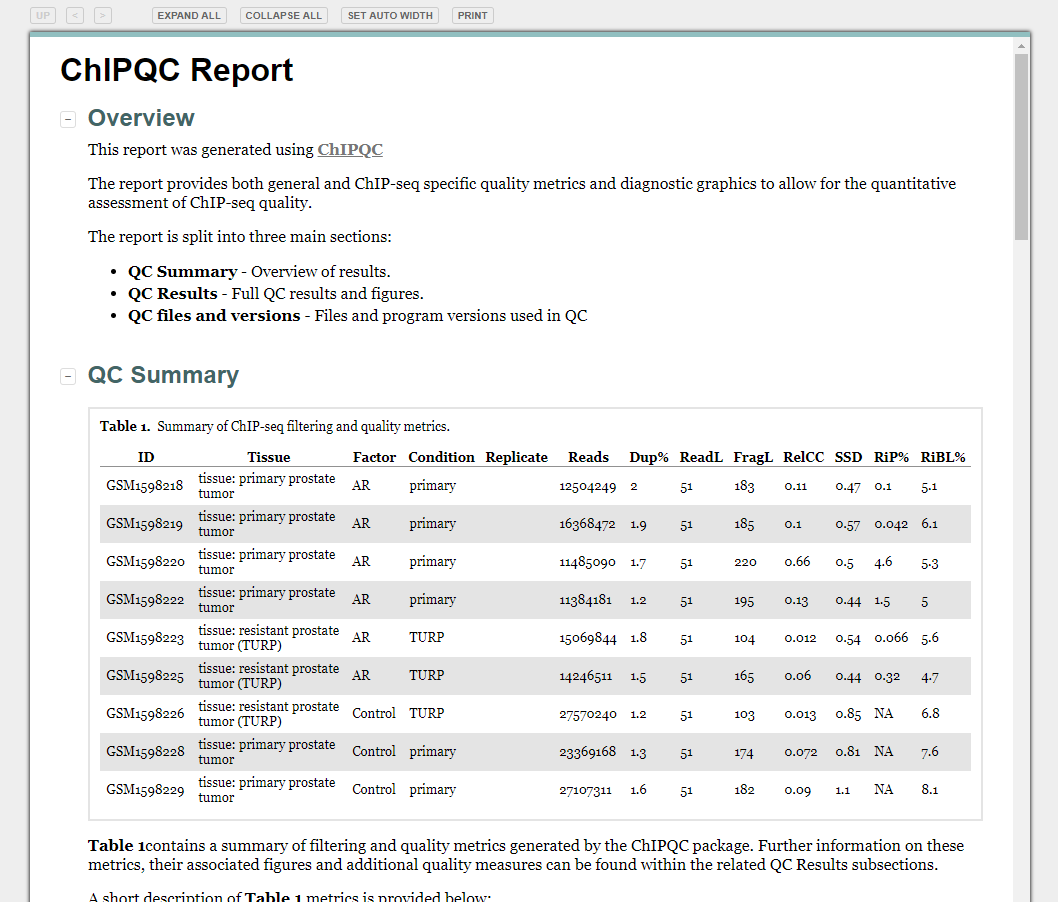
Review: Visualising ChIP-seq data
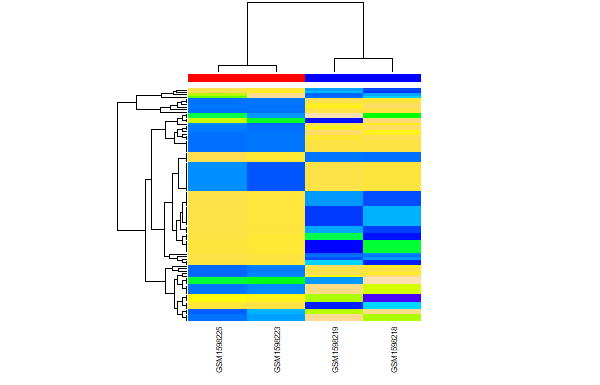
Review: Differential Binding
Review: Enrichment analysis
Moving Ahead
- Time to practice on your own!
- Find datasets to explore at GEO https://www.ncbi.nlm.nih.gov/geo/.
- The data you worked with in this course is available under accession GSE65478.
- Bioconductor website: https://bioconductor.org/
- Bioconductor support: https://support.bioconductor.org/
Well done!
ChIP-seq with Bioconductor in R