Cleaning ChIP-seq data
ChIP-seq with Bioconductor in R

Peter Humburg
Statistician, Macquarie University
Common Problems
Incorrectly mapped reads may produce false peaks.
- Genomic repeats.
Common Problems
Incorrectly mapped reads may produce false peaks.
- Genomic repeats.
- Incomplete reference sequence.
Common Problems
Incorrectly mapped reads may produce false peaks.
- Genomic repeats.
- Incomplete reference sequence.
- Low complexity regions.

Amplification Bias
- DNA fragments extracted from cells are copied multiple times prior to sequencing.
- Not all fragments produce the same number of copies.
- Multiple copies of the same fragment may be sequenced.
- A single DNA fragment may inflate coverage and lead to incorrect peak calls.
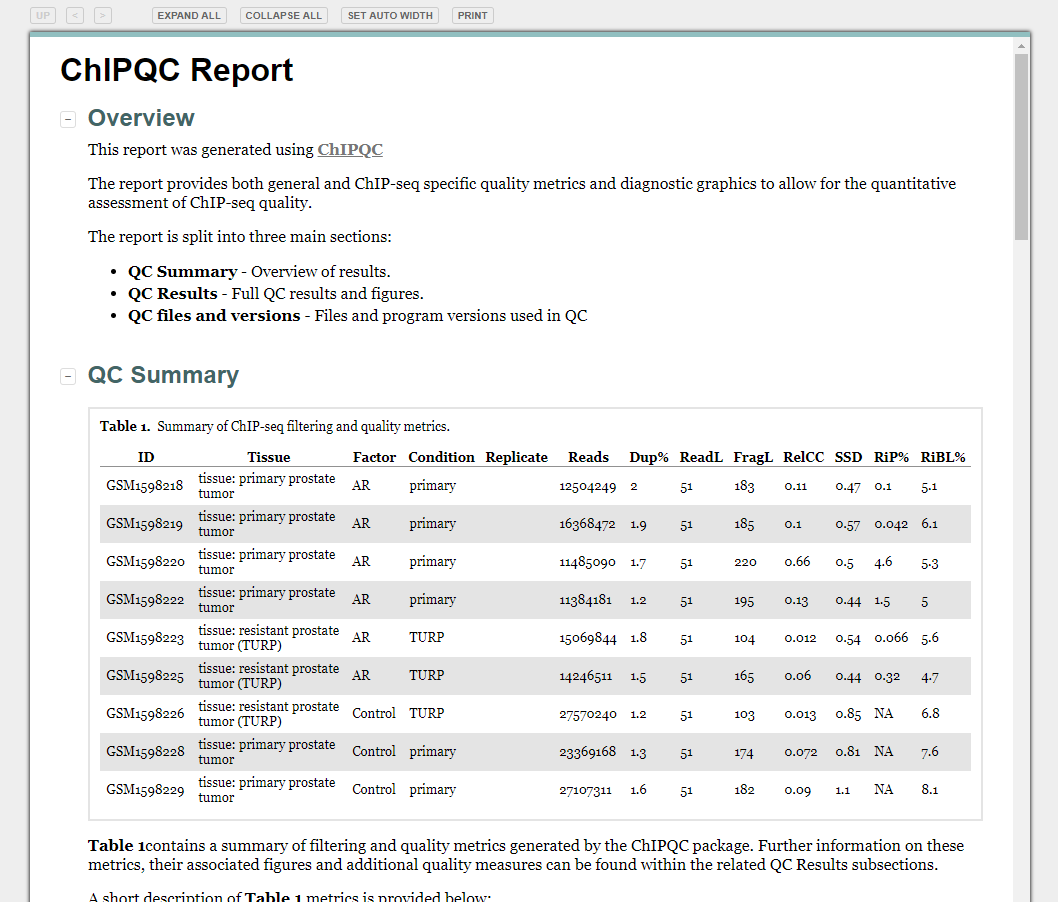
Quality Control Reports
library(ChIPQC)
qc_report <- ChIPQC(experiment="sample_info.csv", annotation="hg19")
ChIPQCreport(qc_report)
Preparing input files
| SampleID | Factor | Condition | Tissue | Treatment | bamReads | Peaks | PeakCaller |
|---|---|---|---|---|---|---|---|
| S1 | AR | primary | primary prostate tumor | gleason score: 3+4=7 | S1.bam | S1.bed | macs |
| S2 | AR | primary | primary prostate tumor | gleason score: 3+4=7 | S2.bam | S2.bed | macs |
| ... | ... | ... | ... | ... | ... | ... | ... |
Cleaning the Data
- Remove duplicate reads.
- Remove reads with multiple hits.
- Remove reads with low mapping quality.
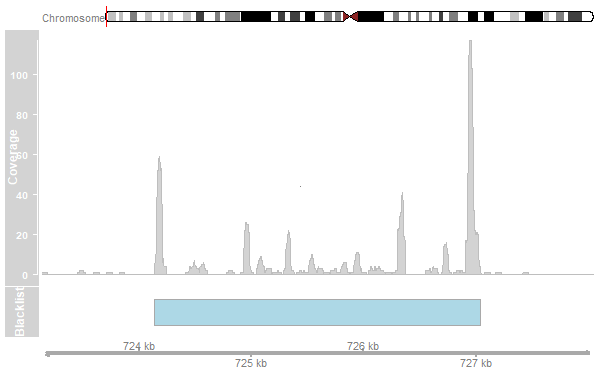
- Remove peaks in blacklisted regions.
Cleaning the Data
- Remove duplicate reads.
- Remove reads with multiple hits.
- Remove reads with low mapping quality.
- Remove peaks in blacklisted regions.
Cleaning the Data
- Remove duplicate reads.
- Remove reads with multiple hits.
- Remove reads with low mapping quality.
- Remove peaks in blacklisted regions.
- Blacklisted regions are available from the ENCODE project.
Let's practice!
ChIP-seq with Bioconductor in R

