Duplicate matches
Joining Data with data.table in R

Scott Ritchie
Postdoctoral Researcher in Systems Genomics
Join key duplicates
# Which bacteria could be found at both sites using any method?
site1_ecology[site2_ecology, on = .(genus)]
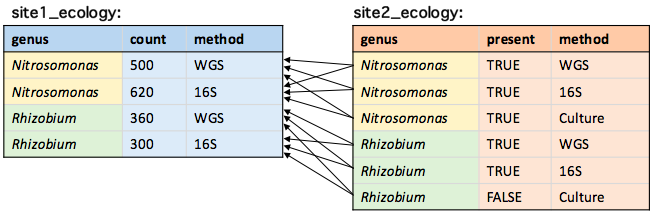
Error from multiplicative matches
site1_ecology[site2_ecology, on = .(genus)]
Error in vecseq(f__, len__, if (allow.cartesian || notjoin ||
!anyDuplicated(f__, :
Join results in 12 rows; more than 10 = nrow(x)+nrow(i). Check for
duplicate key values in i each of which join to the same group in x over
and over again. If that's ok, try by=.EACHI to run j for each group to
avoid the large allocation. If you are sure you wish to proceed, rerun
with allow.cartesian=TRUE. Otherwise, please search for this error message
in the FAQ, Wiki, Stack Overflow and data.table issue tracker for advice.
Allowing multiplicative matches
allow.cartesian = TRUE allows the join to proceed:
# data.table syntax
site1_ecology[site2_ecology, on = .(genus), allow.cartesian = TRUE]
# merge()
merge(site1_ecology, site2_ecology, by = "genus", allow.cartesian = TRUE)
Allowing multiplicative matches
site1_ecology[site2_ecology, on = .(genus), allow.cartesian = TRUE]
genus count method present i.method
1: Nitrosomonas 500 WGS TRUE WGS
2: Nitrosomonas 620 16S TRUE WGS
3: Nitrosomonas 500 WGS TRUE 16S
4: Nitrosomonas 620 16S TRUE 16S
5: Nitrosomonas 500 WGS TRUE Culture
6: Nitrosomonas 620 16S TRUE Culture
7: Rhizobium 360 WGS TRUE WGS
8: Rhizobium 300 16S TRUE WGS
9: Rhizobium 360 WGS TRUE 16S
10: Rhizobium 300 16S TRUE 16S
11: Rhizobium 360 WGS FALSE Culture
12: Rhizobium 300 16S FALSE Culture
Missing values
Missing values (NA) will match all other missing values:
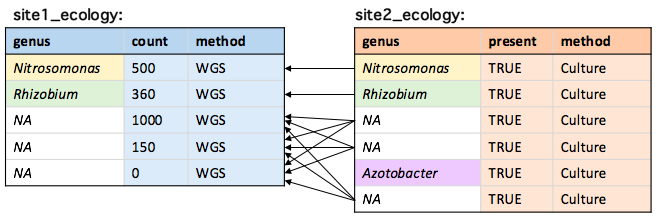
Filtering missing values
!is.na() can be used to filter rows with missing values
site1_ecology <- site1_ecology[!is.na(genus)]
site1_ecology
genus count method
1: Nitrosomonas 500 WGS
2: Rhizobium 360 WGS
site2_ecology <- site2_ecology[!is.na(genus)]
site2_ecology
genus present method
1: Nitrosomonas TRUE Culture
2: Rhizobium TRUE Culture
3: Azotobacter TRUE Culture
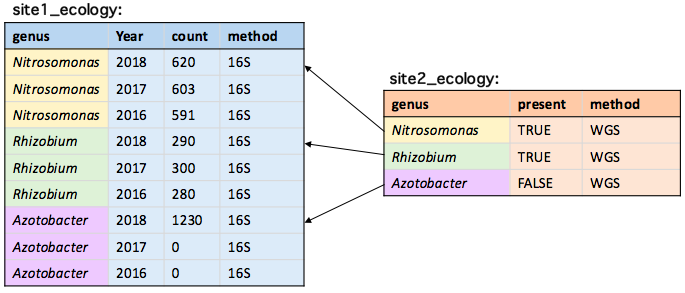
Keeping only the first match
site1_ecology[site2_ecology, on = .(genus), mult = "first"]
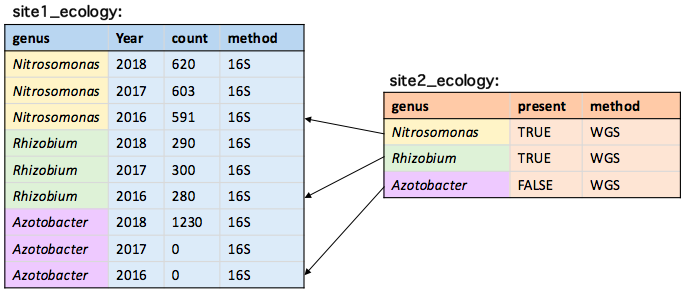
Keeping only the last match
children[parents, on = .(parent = name), mult = "last"]
Identifying and removing duplicates
duplicated(): what rows are duplicates?
unique(): filter a data.table to just unique rows
The duplicated() function
Using values in all columns:
duplicated(site1_ecology)
FALSE FALSE FALSE FALSE
Using values in a subset of columns:
duplicated(site1_ecology,
by = "genus")
FALSE TRUE FALSE TRUE
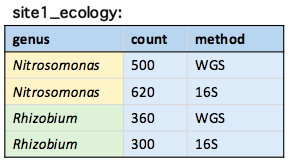
The unique() function
unique(site1_ecology, by = "genus")
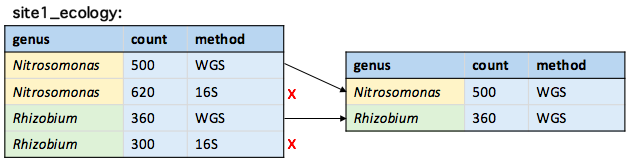
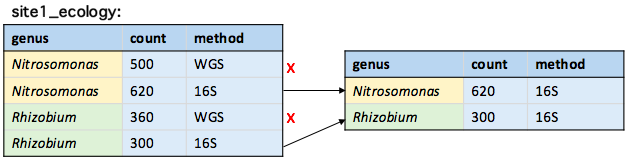
Changing the search order
fromLast = TRUE changes the direction of the search to start from the last row
duplicated(site1_ecology, by = "genus", fromLast = TRUE)
TRUE FALSE TRUE FALSE
unique(site1_ecology, by = "genus", fromLast = TRUE)
Let's practice!
Joining Data with data.table in R

