Point charts
Visualization Best Practices in R

Nick Strayer
Instructor
When a bar chart isn't ideal
- Not a quantity
- Non-linear transformations

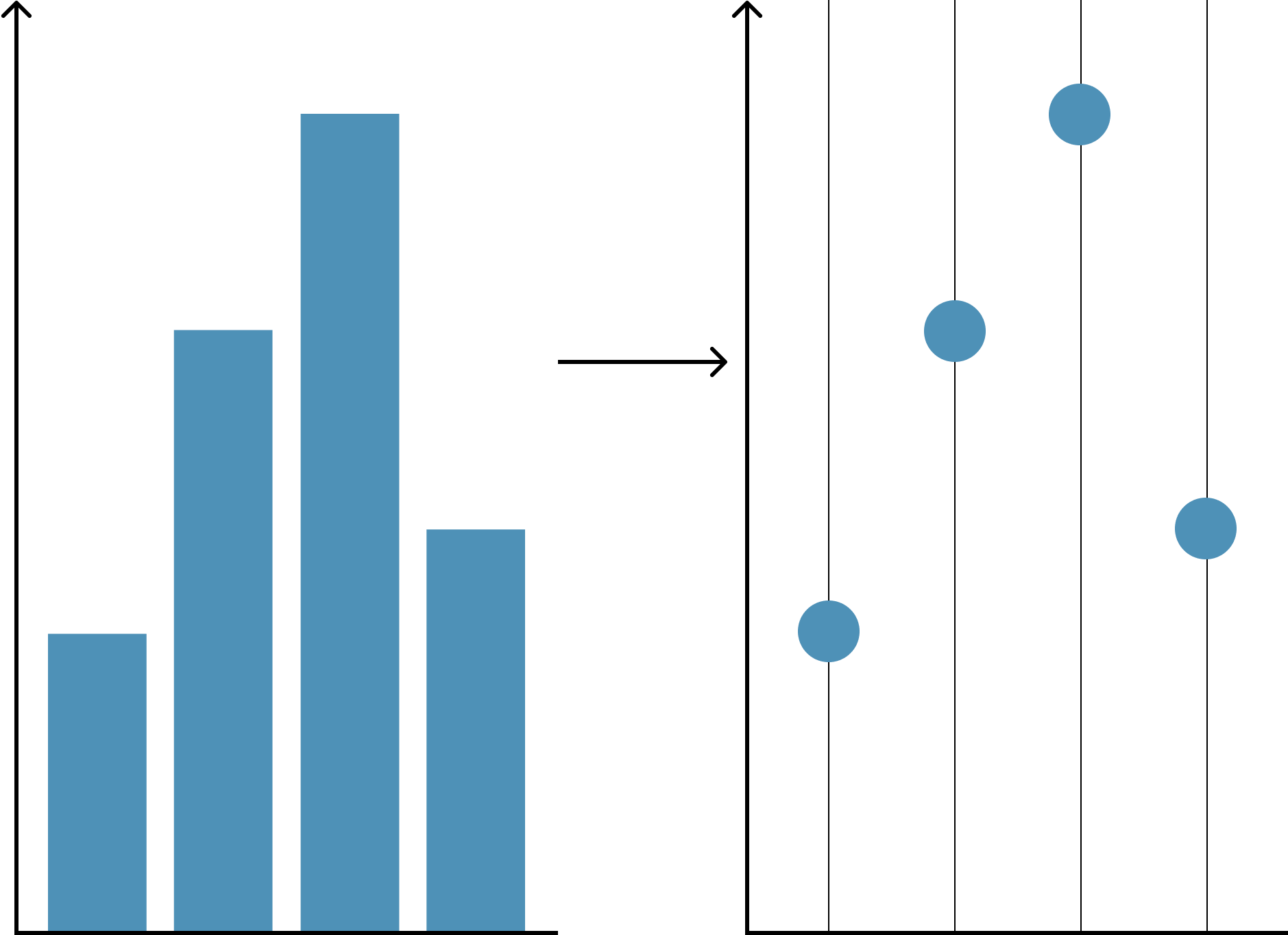
Point charts
- Simply replace bar with a point
- Sometimes called point charts or dot plots
Benefits of point charts
- High precision
- Efficient representation
- Simple
Data for lesson
- Working with a subset of WHO data
- Countries are an 'interesting' subset -- let's see if we can find out why
interestingCountries <- c("NGA", "SDN", "FRA", "NPL", "MYS", "TZA", "YEM", "UKR", "BGD", "VNM")
who_subset <- who_disease %>%
filter(
countryCode %in% interestingCountries,
disease == 'measles',
year %in% c(2006, 2016)) %>%
mutate(year = paste0('cases_', year)) %>%
arrange(year, region) %>%
pivot_wider(names_from = year, values_from = cases)
who_subset
who_subset
# A tibble: 10 x 6
region countryCode country disease cases_2006 cases_2016
<chr> <chr> <chr> <chr> <dbl> <dbl>
1 AFR NGA Nigeria measles 704 17136
2 AFR TZA Tanzania measles 2362 33
3 EMR SDN Sudan (the) measles 228 1767
4 EMR YEM Yemen measles 8079 143
5 EUR FRA France measles 40 79
6 EUR UKR Ukraine measles 42724 102
7 SEAR BGD Bangladesh measles 6192 972
8 SEAR NPL Nepal measles 2838 1269
9 WPR MYS Malaysia measles 564 1569
10 WPR VNM Viet Nam measles 1978 46
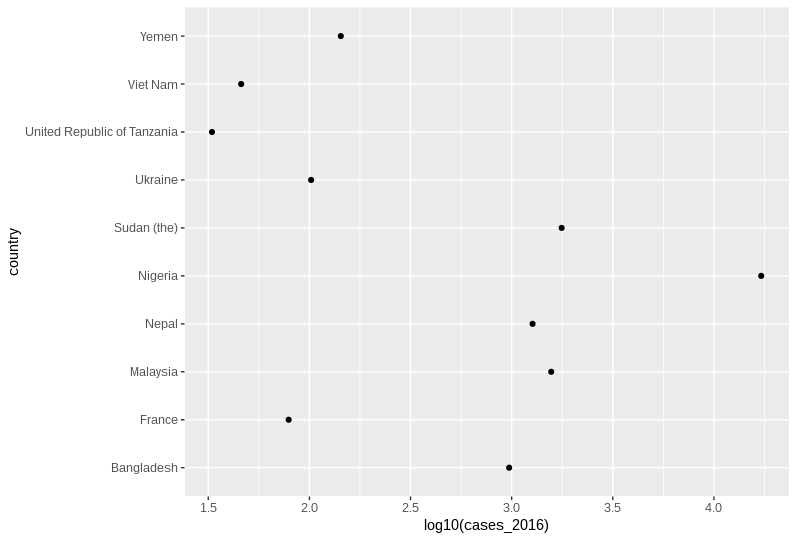
Code for point charts
geom_point()with one categorical and one numerical axis
who_subset %>%
# We log transform our values here so bars are not appropriate
ggplot(aes(y = country, x = log10(cases_2016))) +
# Simple geom_point()
geom_point()
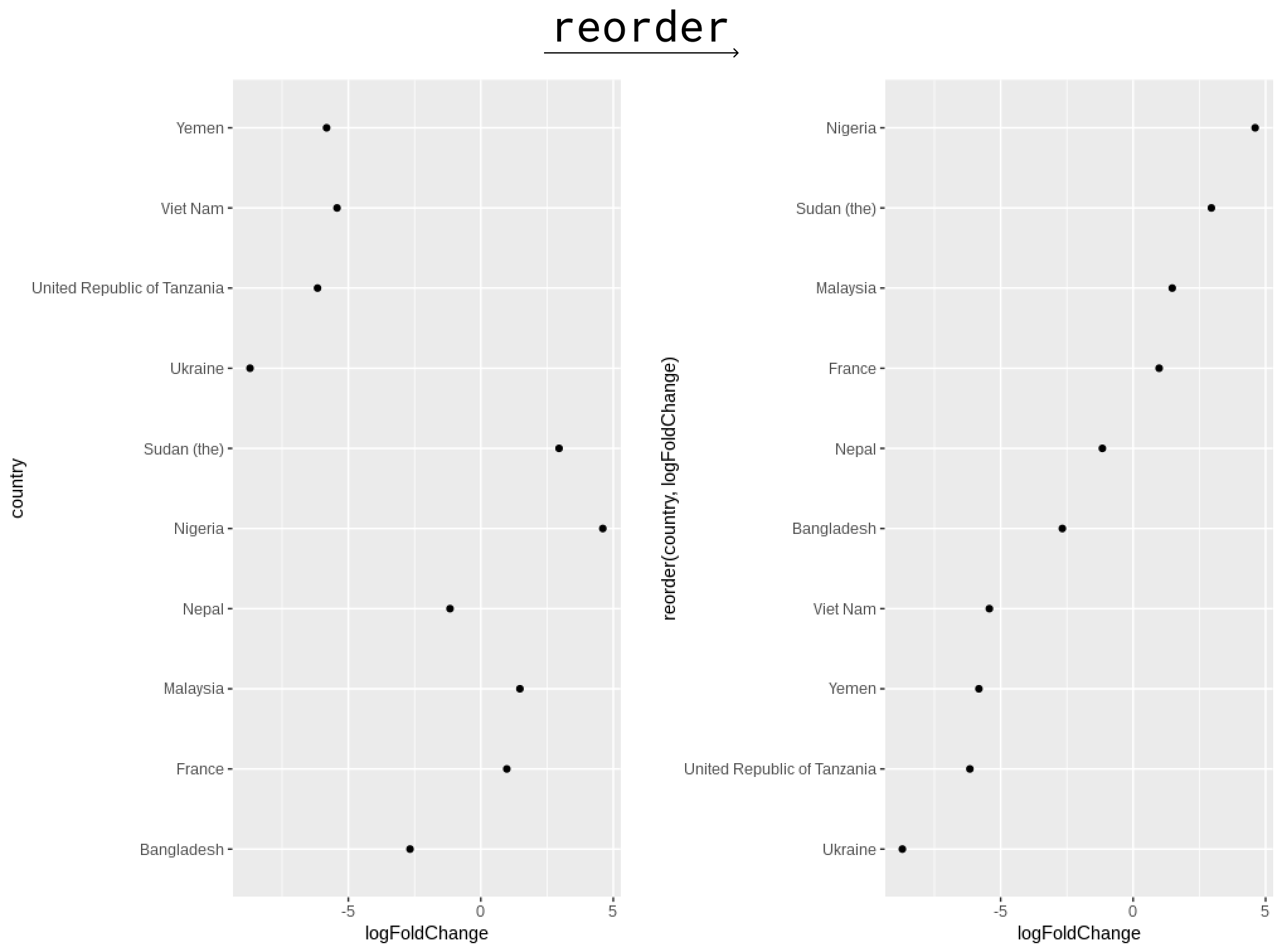
Ordering your point charts
- Ordering can vastly help legibility
- Use the
reorder()function in the aesthetic assignment
who_subset %>%
# calculate the log fold change between 2016 and 2006
mutate(logFoldChange = log2(cases_2016/cases_2006)) %>%
ggplot(aes(x = logFoldChange, y = reorder(country, logFoldChange))) +
geom_point()
Let's practice!
Visualization Best Practices in R

