Multiple and parallel sequence quality assessment
Introduction to Bioconductor in R

Paula Andrea Martinez, PhD.
Data Scientist
Rqc
library(Rqc)
- Uses Bioconductor packages that you have already used:
Biostrings,IRanges,methods,S4vectors
- New packages to discover in the following Bioconductor courses:
Rsamtools,GenomicAlignments,GenomicFiles,BiocParallel
- CRAN packages:
Knitr,dplyr,markdown,ggplot2,digest,shiny, andRcpp
rqcQA
library(Rqc) files <- # get the full path of the files you want to assess qaRqc <- rqcQA(files)# exploring qaRqc class(qaRqc) # "list" names(qaRqc) # name of the input files# for each file qaRqc[1] # the class of the results is RqcResultSet
rqcQA arguments
library(Rqc) # get the path of the files you want to assess files <- "data/seq1.fq" "data/seq2.fq" "data/seq3.fq" "data/se4.fq" qaRqc <- rqcQA(files, workers = 4)# sample of sequences set.seed(1111) qaRqc_sample <- rqcQA(files, workers = 4, sample = TRUE, n = 500))# paired-end files pfiles <- "data/seq_11.fq" "data/seq1_2.fq" "data/seq2_1.fq" "data/seq2_2.fq" qaRqc_paired <- rqcQA(pfiles, workers = 4, pair = c(1, 1, 2, 2)))
rqcReport and rqcResultSet
# create a report reportFile <- rqcReport(qaRqc, templateFile = "myReport.Rmd")browseURL(reportFile)#The class of qaRqc is rqcResultSet methods(class = "RqcResultSet")
perFileInformation
qaRqc <- rqcQA(files, workers = 4))
perFileInformation(qaRqc)
filename pair format group reads total.reads path
SRR7760274.fastq 1 FASTQ None 1e+06 2404795 ./data
SRR7760275.fastq 2 FASTQ None 1e+06 1508139 ./data
SRR7760276.fastq 3 FASTQ None 1e+06 1950463 ./data
SRR7760277.fastq 4 FASTQ None 1e+06 2629588 ./data
Plot functions
| rqc Plot functions | rqc Plot functions |
|---|---|
rqcCycleAverageQualityPcaPlot() |
rqcGroupCycleAverageQualityPlot() |
rqcCycleAverageQualityPlot() |
rqcReadQualityBoxPlot() |
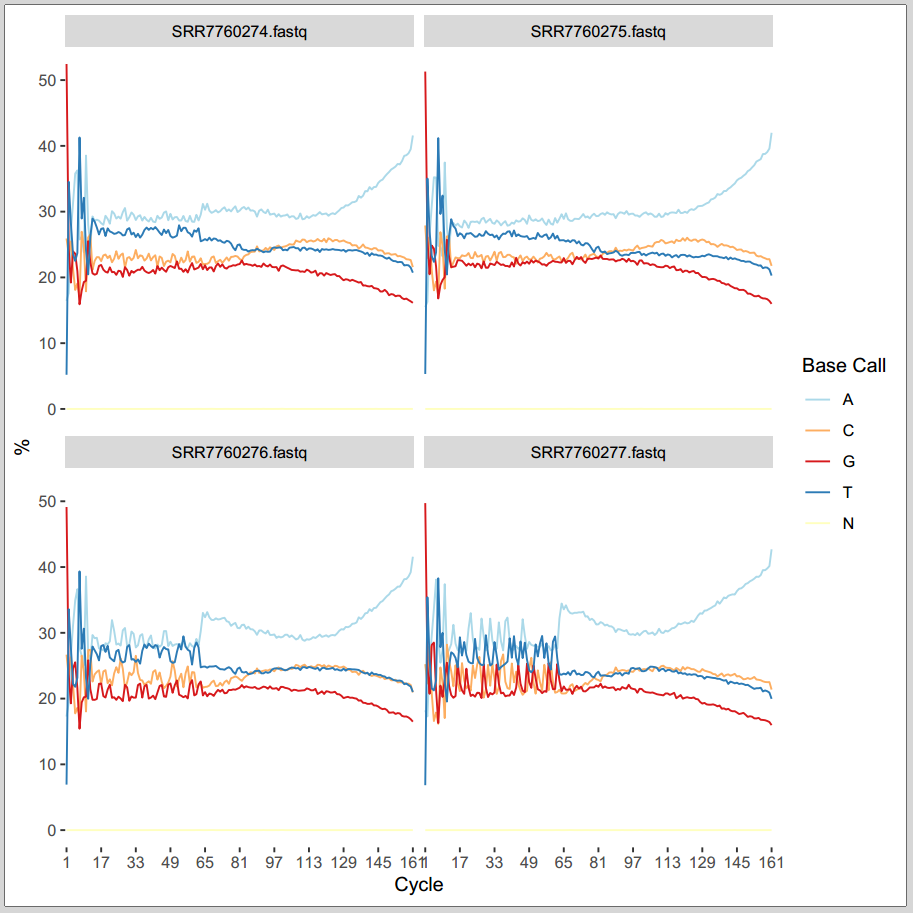
rqcCycleBaseCallsLinePlot() |
rqcReadQualityPlot() |
rqcCycleBaseCallsPlot() |
rqcReadWidthPlot() |
rqcCycleGCPlot() |
rqcReadFrequencyPlot() |
rqcCycleQualityBoxPlot() |
rqcCycleQualityPlot() |

You are ready!
Introduction to Bioconductor in R

