Why are we interested in patterns?
Introduction to Bioconductor in R

James Chapman
Curriculum Manager, DataCamp

What can we find with patterns?
- Gene start
- Protein end
- Regions that enhance or silence gene expression
- Conserved regions between organisms
- Genetic variation
Pattern matching
Biostringsprovides functions for pattern matchingmatchPattern(pattern, subject)- 1 string to 1 string
vmatchPattern(pattern, subject)- 1 set of strings to 1 string
- 1 string to a set of strings
Palindromes

findPalindromes() # find palindromic regions in a single sequence
Not new biology
The Genetic code was first described by Nirenberg in 1963 On the coding of genetic information Nirenberg, Marshall et al. Cold Spring Harb Symp Quant Biol 1963, 28
How translation might differ according to the reading frame, was first described by Streisinger in 1966 Frameshift Mutations and the Genetic Code Streisinger, George et al. Cold Spring Harb Symp Quant Biol 1966, 31: 77-84
# Original dna sequence
[1] 30 ACATGGGCCTACCATGGGAGCTACGAAGCC
# 6 possible reading frames, DNAStringSet
[1] 30 ACATGGGCCTACCATGGGAGCTACGAAGCC + 1
[2] 30 GGCTTCGTAGCTCCCATGGTAGGCCCATGT - 1
[3] 29 CATGGGCCTACCATGGGAGCTACGAAGCC + 2
[4] 29 GCTTCGTAGCTCCCATGGTAGGCCCATGT - 2
[5] 28 ATGGGCCTACCATGGGAGCTACGAAGCC + 3
[6] 28 CTTCGTAGCTCCCATGGTAGGCCCATGT - 3
# 6 possible translations, AAStringSet
[1] 10 TWAYHGSYEA + 1
[2] 10 GFVAPMVGPC - 1
[3] 9 HGPTMGATK + 2
[4] 9 AS*LPW*AH - 2
[5] 9 MGLPWELRS + 3
[6] 9 LRSSHGRPM - 3
Conserved regions in the Zika virus
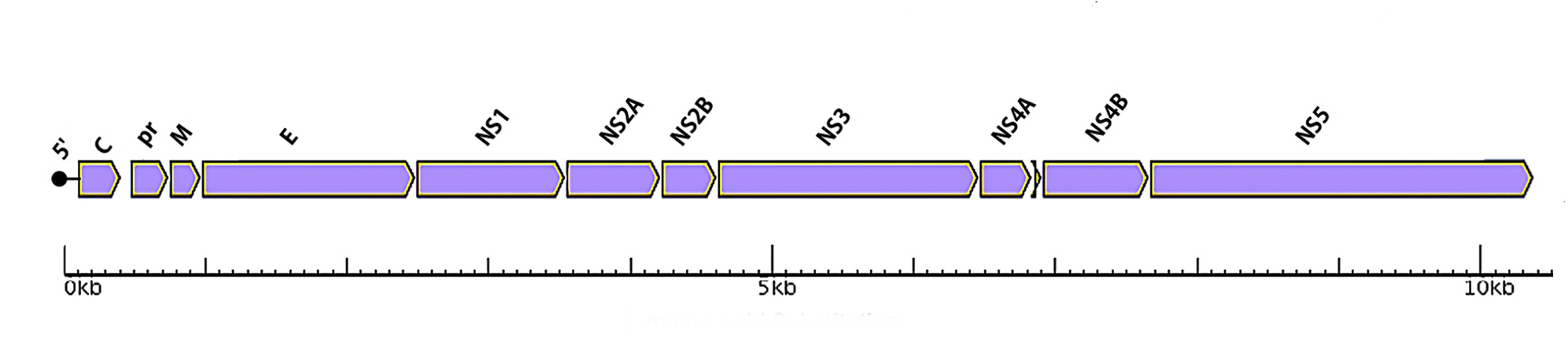 Adapted figure From Mosquitos to Humans: Genetic Evolution of Zika Virus
Wang, Lulan et al. Cell Host & Microbe 2016, Vol 19 5: 561-565
Adapted figure From Mosquitos to Humans: Genetic Evolution of Zika Virus
Wang, Lulan et al. Cell Host & Microbe 2016, Vol 19 5: 561-565
Facts
- The Zika Virus has a positive strand genome
- It lives in humans, monkeys, and mosquitoes
- The Flaviviruses family and share 11 conserved proteins
Let's practice finding patterns!
Introduction to Bioconductor in R

