Introduction to Biostrings
Introduction to Bioconductor in R

James Chapman
Curriculum Manager, DataCamp
Biostrings
- Algorithms for fast manipulation of sequences
- Many Bioconductor packages are dependent on
Biostrings
BiocManager::install("Biostrings")
Biological string containers
- Biostrings → Memory efficient to store and manipulate sequence of characters
- Containers that can be inherited
For example:
- The BString class comes from big string
Strings vs. Sets
- XString to store a single sequence
- BString for any string
- DNAString for DNA
- RNAString for RNA
- AAString for amino acids
- XStringSet for many sequences
- BStringSet
- DNAStringSet
- RNAStringSet
- AAStringSet
showClass()
showClass("XString")
Virtual Class "XString" [package "Biostrings"]
Slots:
Name: shared offset length elementMetadata metadata
Class: SharedRaw integer integer DataFrame_OR_NULL list
Extends:
Class "XRaw", directly
Class "XVector", by class "XRaw", distance 2
Class "Vector", by class "XRaw", distance 3
Class "Annotated", by class "XRaw", distance 4
Class "vector_OR_Vector", by class "XRaw", distance 4
Known Subclasses: "BString", "DNAString", "RNAString", "AAString"
Biostring alphabets
DNA_BASES # 4 DNA basesRNA_BASES # 4 RNA bases
"A" "C" "G" "T""A" "C" "G" "U"
AA_STANDARD # 20 Amino acids
"A" "R" "N" "D" "C" "Q" "E" "G" "H" "I" "L" "K" "M" "F" "P" "S" "T" "W" "Y" "V"
DNA_ALPHABET # contains IUPAC_CODE_MAP
RNA_ALPHABET # contains IUPAC_CODE_MAP
AA_ALPHABET # contains AMINO_ACID_CODE
1 For more information IUPAC DNA codes http://genome.ucsc.edu/goldenPath/help/iupac.html
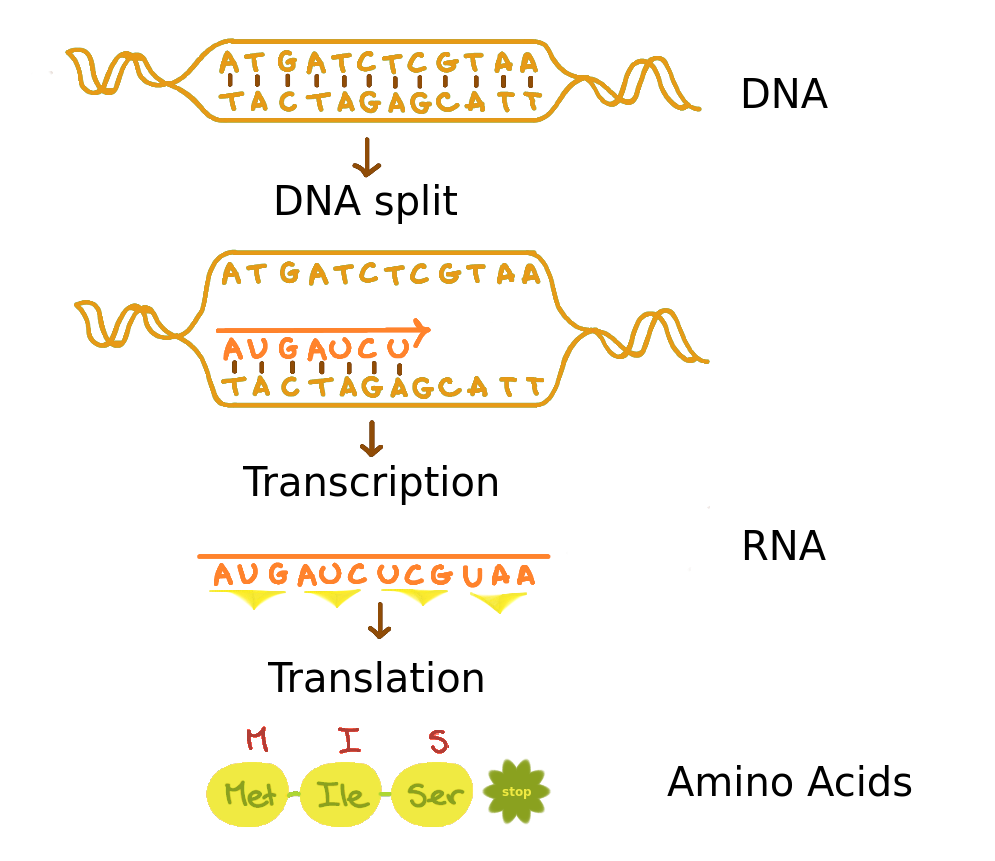
Transcription DNA to RNA
# DNA single string
dna_seq <- DNAString("ATGATCTCGTAA")
dna_seq
12-letter DNAString object
seq: ATGATCTCGTAA
# Transcription DNA to RNA string
rna_seq <- RNAString(dna_seq)
rna_seq
12-letter RNAString object
seq: AUGAUCUCGUAA
Translation RNA to amino acids
rna_seq
12-letter RNAString object
seq: AUGAUCUCGUAA
# Translation RNA to AA
aa_seq <- translate(rna_seq)
aa_seq
Three RNA bases form one AA: AUG = M, AUC = I, UCG = S, UAA = *
4-letter AAString object
seq: MIS*
Shortcut translate DNA to amino acids
dna_seq
12-letter DNAString object
seq: ATGATCTCGTAA
# translate() also goes directly from DNA to AA
translate(dna_seq)
4-letter AAString object
seq: MIS*
The Zika virus
Let's practice with the Zika virus!
Introduction to Bioconductor in R

