Sequence handling
Introduction to Bioconductor in R

James Chapman
Curriculum Manager, DataCamp
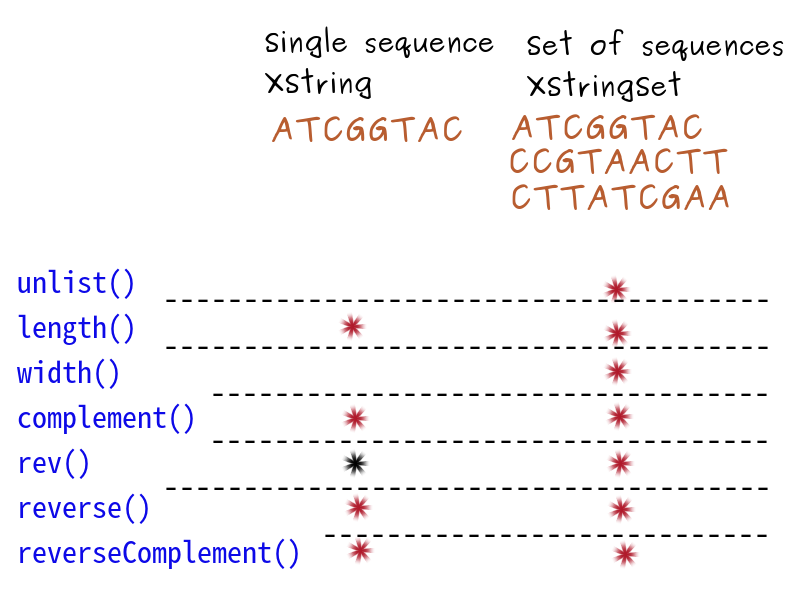
Single vs. Set
- XString to store a single sequence
- BString for any string
- DNAString for DNA
- RNAString for RNA
- AAString for amino acids
- XStringSet for many sequences
- BStringSet
- DNAStringSet
- RNAStringSet
- AAStringSet
Create a StringSet and collate it
# Read the sequence as a set zikaVirus <- readDNAStringSet("data/zika.fa")length(zikaVirus) # the set contains only one sequence width(zikaVirus) # and width 10794 bases
1
10794
# Collate the sequence zikaVirus_seq <- unlist(zikaVirus)length(zikaVirus_seq)width(zikaVirus_seq)
10794Error in (function (classes, fdef, mtable) : unable to find an inherited method for function ‘width’ for signature ‘"DNAString"’
From a single sequence to a set
# to create a new set from a single sequence
zikaSet <- DNAStringSet(zikaVirus_seq, start = c(1, 101, 201), end = c(100, 200, 300))
zikaSet
DNAStringSet object of length 3:
width seq
[1] 100 AGTTGTTGATCTGTGTGAGTCAGACTGCGACAGTTCGAGTCTGAAG...AACAACAGTATCAACAGGTTTAATTTGGATTTGGAAACGAGAGTTT
[2] 100 CTGGTCATGAAAAACCCCAAAGAAGAAATCCGGAGGATCCGGATTG...CTAAAACGCGGAGTAGCCCGTGTAAACCCCTTGGGAGGTTTGAAGA
[3] 100 GGTTGCCAGCCGGACTTCTGCTGGGTCATGGACCCATCAGAATGGT...TACTAGCCTTTTTGAGATTTACAGCAATCAAGCCATCACTGGGCCT
length(zikaSet)
width(zikaSet)
3
100 100 100
Complement sequence

a_seq <- DNAString("ATGATCTCGTAA")
a_seq
12-letter DNAString object
seq: ATGATCTCGTAA
complement(a_seq)
12-letter DNAString object
seq: TACTAGAGCATT
Rev a sequence
zikaShortSet
DNAStringSet instance of length 2
width seq names
[1] 18 AGTTGTTGATCTGTGTGA seq1
[2] 18 CTGGTCATGAAAAACCCC seq2
rev(zikaShortSet)
A DNAStringSet instance of length 2
width seq names
[1] 18 CTGGTCATGAAAAACCCC seq2
[2] 18 AGTTGTTGATCTGTGTGA seq1
Reverse a sequence
zikaShortSet
A DNAStringSet instance of length 2
width seq names
[1] 18 AGTTGTTGATCTGTGTGA seq1
[2] 18 CTGGTCATGAAAAACCCC seq2
reverse(zikaShortSet)
A DNAStringSet instance of length 2
width seq names
[1] 18 AGTGTGTCTAGTTGTTGA seq1
[2] 18 CCCCAAAAAGTACTGGTC seq2
Reverse complement
# Original rna_seq sequence
8-letter RNAString object
seq: AGUUGUUG
reverseComplement(rna_seq)
8-letter RNAString object
seq: CAACAACU
# Using two functions together
reverse(complement(rna_seq))
8-letter RNAString object
seq: CAACAACU
Let's practice sequence handling!
Introduction to Bioconductor in R

