Introducing biology of genomic datasets
Introduction to Bioconductor in R

James Chapman
Curriculum Manager, DataCamp
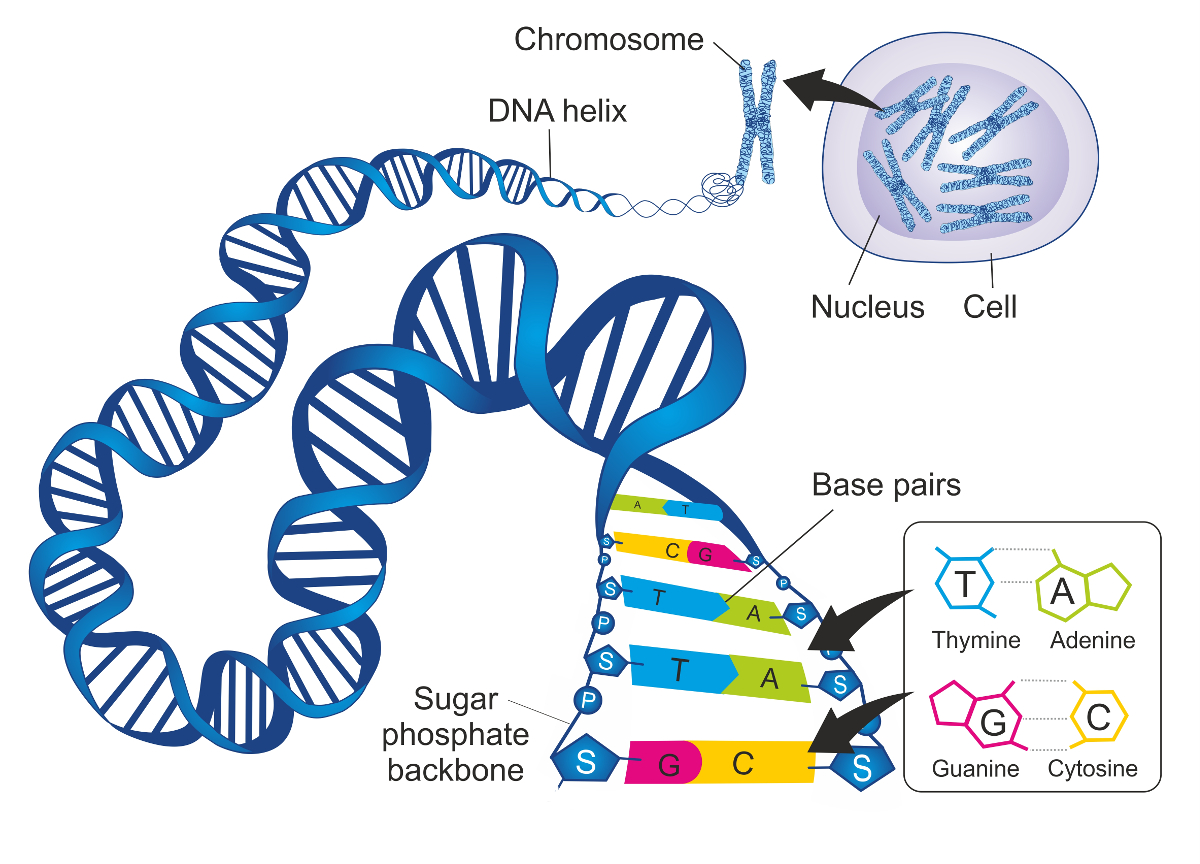
Genome elements
- Genetic information DNA alphabet
- A set of chromosomes (highly variable number)
- Genes (carry heredity instructions)
- coding and non-coding
- Proteins (responsible for specific functions)
- DNA-to-RNA (transcription)
- RNA-to-protein (translation)
Yeast
- A single cell microorganism
- The fungus that people love ♥
- Used for fermentation: beer, bread, kefir, kombucha, bioremediation, etc.
- Name: Saccharomyces cerevisiae or S. cerevisiae

BSgenome annotation package
# Load the package and store data into yeast library(BSgenome.Scerevisiae.UCSC.sacCer3) yeast <- BSgenome.Scerevisiae.UCSC.sacCer3# Other available genomes available.genomes()
"BSgenome.Alyrata.JGI.v1"
"BSgenome.Amellifera.BeeBase.assembly4"
"BSgenome.Amellifera.NCBI.AmelHAv3.1"
"BSgenome.Amellifera.UCSC.apiMel2"
"BSgenome.Amellifera.UCSC.apiMel2.masked"
...
length(yeast)
17
names(yeast)
"chrI" "chrII" "chrIII" "chrIV" "chrV" "chrVI" "chrVII"
"chrVIII" "chrIX" "chrX" "chrXI" "chrXII" "chrXIII" "chrXIV"
"chrXV" "chrXVI" "chrM"
seqlengths(yeast)
chrI chrII chrIII chrIV chrV chrVI chrVII chrVIII chrIX chrX
230218 813184 316620 1531933 576874 270161 1090940 562643 439888 745751
chrXI chrXII chrXIII chrXIV chrXV chrXVI chrM
666816 1078177 924431 784333 1091291 948066 85779
Get sequences
getSeq(): S4 method for BSgenome
# Select entire genomic sequence getSeq(yeast)# Select sequence from chromosome M getSeq(yeast, "chrM")# Select first 10 base pairs getSeq(yeast, end = 10)
Let's practice!
Introduction to Bioconductor in R

