Bayesian regression in RJAGS
Bayesian Modeling with RJAGS

Alicia Johnson
Associate Professor, Macalester College
Bayesian regression model
$Y_i$ = weight of adult $i$ (kg)
$X_i$ = height of adult $i$ (cm)
$\;$
Model
$Y_i \sim N(m_i, s^2)$
$m_i = a + b X_i$
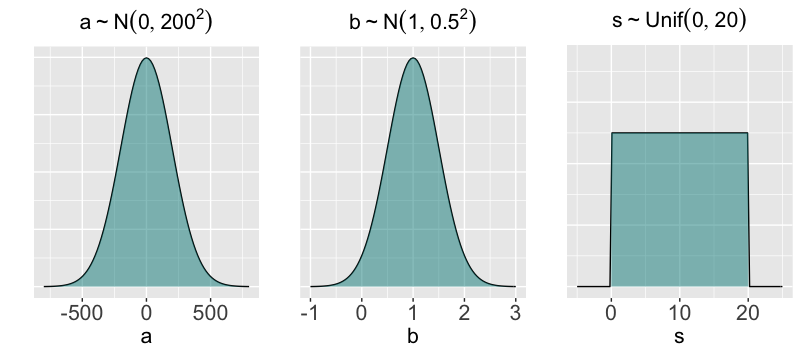
$a \sim N(0, 200^2)$
$b \sim N(1, 0.5^2)$
$s \sim \text{Unif}(0, 20)$
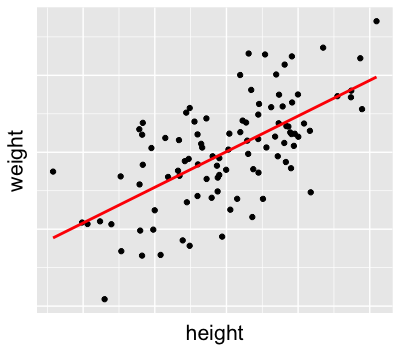
Prior insight
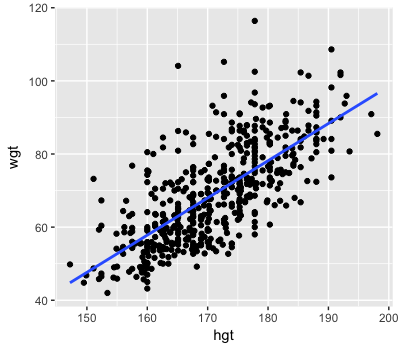
Insight from the observed weight & height data
$Y_i \sim N(m_i, s^2)$
$m_i = a + b X_i$
wt_mod <- lm(wgt ~ hgt, bdims)
coef(wt_mod)
(Intercept) hgt
-105.011254 1.017617
summary(wt_mod)$sigma
9.30804
DEFINE the regression model
weight_model <- "model{
# Likelihood model for Y[i]
# Prior models for a, b, s
}"
DEFINE the regression model
weight_model <- "model{
# Likelihood model for Y[i]
for(i in 1:length(Y)) {
}
# Prior models for a, b, s
}"
- $Y_i \sim N(m_i, s^2)$ for $i$ from 1 to 507
DEFINE the regression model
weight_model <- "model{
# Likelihood model for Y[i]
for(i in 1:length(Y)) {
Y[i] ~ dnorm(m[i], s^(-2))
}
# Prior models for a, b, s
}"
- $Y_i \sim N(m_i, s^2)$ for $i$ from 1 to 507
DEFINE the regression model
weight_model <- "model{
# Likelihood model for Y[i]
for(i in 1:length(Y)) {
Y[i] ~ dnorm(m[i], s^(-2))
m[i] <- a + b * X[i]
}
# Prior models for a, b, s
}"
- $Y_i \sim N(m_i, s^2)$ for $i$ from 1 to 507
- $m_i = a + b X_i$
- NOTE: use
<-not~
- NOTE: use
DEFINE the regression model
weight_model <- "model{
# Likelihood model for Y[i]
for(i in 1:length(Y)) {
Y[i] ~ dnorm(m[i], s^(-2))
m[i] <- a + b * X[i]
}
# Prior models for a, b, s
a ~ dnorm(0, 200^(-2))
b ~ dnorm(1, 0.5^(-2))
s ~ dunif(0, 20)
}"
- $Y_i \sim N(m_i, s^2)$ for $i$ from 1 to 507
$m_i = a + b X_i$
- NOTE: use
<-not~
- NOTE: use
$a \sim N(0, 200^2)$
- $b \sim N(1, 0.5^2)$
- $s \sim \text{Unif}(0, 20)$
COMPILE the regression model
# COMPILE the model weight_jags <- jags.model(textConnection(weight_model), data = list(X = bdims$hgt, Y = bdims$wgt), inits = list(.RNG.name = "base::Wichmann-Hill", .RNG.seed = 2018))dim(bdims)
507 25
head(bdims$hgt)head(bdims$wgt)
174.0 175.3 193.5 186.5 187.2 181.565.6 71.8 80.7 72.6 78.8 74.8
SIMULATE the regression model
# COMPILE the model
weight_jags <- jags.model(textConnection(weight_model),
data = list(X = bdims$hgt, Y = bdims$wgt),
inits = list(.RNG.name = "base::Wichmann-Hill",
.RNG.seed = 2018))
# SIMULATE the posterior
weight_sim <- coda.samples(model = weight_jags,
variable.names = c("a", "b", "s"),
n.iter = 10000)
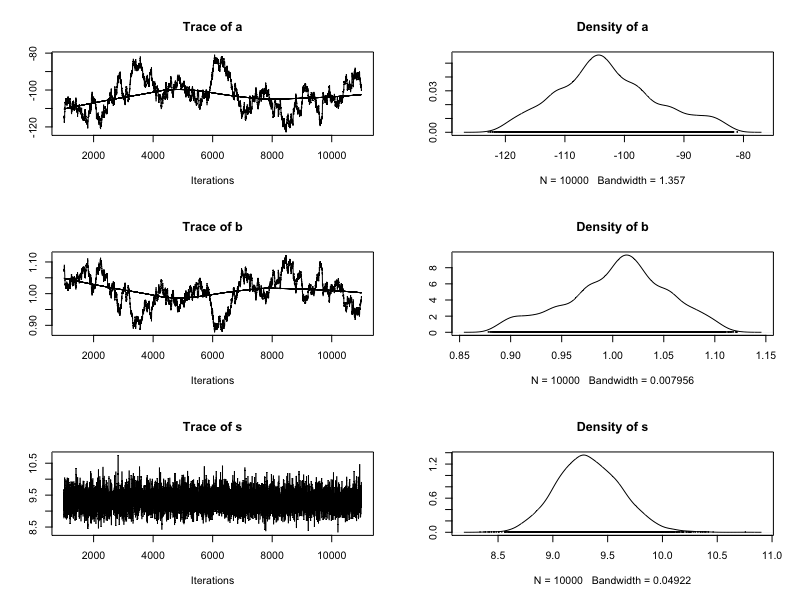
Addressing Markov chain instability
Standardize the height predictor (subtract the mean and divide by the standard deviation)
Increase chain length
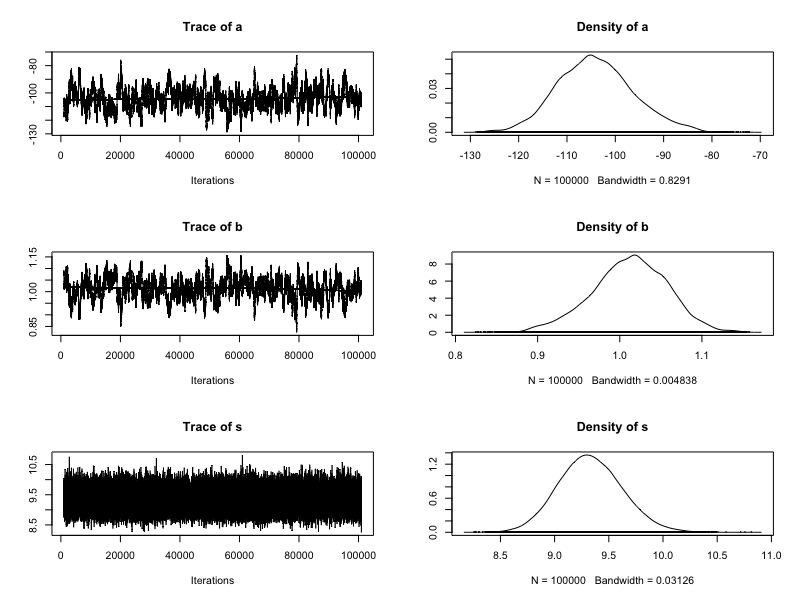
Posterior insights
Let's practice!
Bayesian Modeling with RJAGS

